Integrate lanes from all batches of EB pilot (Human data only, no doublets
Last updated: 2020-08-10
Checks: 7 0
Knit directory: Embryoid_Body_Pilot_Workflowr/analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20200804) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version f50ebd3. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: output/.Rhistory
Untracked files:
Untracked: analysis/child/
Untracked: code/EB.getHumanMetadata.Rmd
Untracked: figure/
Untracked: output/mergedObjects/
Untracked: output/sampleQCrds/
Unstaged changes:
Modified: .Rprofile
Modified: .gitattributes
Modified: .gitignore
Modified: Embryoid_Body_Pilot_Workflowr.Rproj
Modified: README.md
Modified: _workflowr.yml
Modified: analysis/_site.yml
Modified: analysis/about.Rmd
Modified: analysis/index.Rmd
Modified: analysis/license.Rmd
Modified: code/README.md
Modified: data/README.md
Modified: output/README.md
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/IntegrateAnalysis.afterFilter.HarmonyBatch.Rmd) and HTML (docs/IntegrateAnalysis.afterFilter.HarmonyBatch.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | f50ebd3 | KLRhodes | 2020-08-10 | wflow_publish("analysis/Integrate*") |
| html | 421a225 | KLRhodes | 2020-08-10 | Build site. |
| Rmd | bc8ec6f | KLRhodes | 2020-08-10 | cleaning various versions of merging/intCurrent working directory |
library(Seurat)
library(harmony)
library(ggplot2)
library(DataCombine)
library(here)
library(RColorBrewer)
options(future.globals.maxSize= 15000*1024^2) #allow global exceeding 4GbRead in the files, add metadata, and create an object list
filelist<-list.files(here::here('output/sampleQCrds/'), full.names = T)
objectlist<- list()
for (i in 1:length(filelist)){
rds<- readRDS(filelist[i])
objectlist[i]<- rds
}Warning in `[<-`(`*tmp*`, i, value = rds): implicit list embedding of S4 objects
is deprecated
Warning in `[<-`(`*tmp*`, i, value = rds): implicit list embedding of S4 objects
is deprecated
Warning in `[<-`(`*tmp*`, i, value = rds): implicit list embedding of S4 objects
is deprecated
Warning in `[<-`(`*tmp*`, i, value = rds): implicit list embedding of S4 objects
is deprecated
Warning in `[<-`(`*tmp*`, i, value = rds): implicit list embedding of S4 objects
is deprecated
Warning in `[<-`(`*tmp*`, i, value = rds): implicit list embedding of S4 objects
is deprecated
Warning in `[<-`(`*tmp*`, i, value = rds): implicit list embedding of S4 objects
is deprecated
Warning in `[<-`(`*tmp*`, i, value = rds): implicit list embedding of S4 objects
is deprecated
Warning in `[<-`(`*tmp*`, i, value = rds): implicit list embedding of S4 objects
is deprecated
Warning in `[<-`(`*tmp*`, i, value = rds): implicit list embedding of S4 objects
is deprecated
Warning in `[<-`(`*tmp*`, i, value = rds): implicit list embedding of S4 objects
is deprecated
Warning in `[<-`(`*tmp*`, i, value = rds): implicit list embedding of S4 objects
is deprecated
Warning in `[<-`(`*tmp*`, i, value = rds): implicit list embedding of S4 objects
is deprecated
Warning in `[<-`(`*tmp*`, i, value = rds): implicit list embedding of S4 objects
is deprecated
Warning in `[<-`(`*tmp*`, i, value = rds): implicit list embedding of S4 objects
is deprecated
Warning in `[<-`(`*tmp*`, i, value = rds): implicit list embedding of S4 objects
is deprecatedcreate a merged seurat object
ids<-substr(basename(filelist),1,12)
merged<- merge(objectlist[[1]], c(objectlist[[2]], objectlist[[3]],objectlist[[4]],objectlist[[5]],objectlist[[6]],objectlist[[7]],objectlist[[8]],objectlist[[9]],objectlist[[10]],objectlist[[11]],objectlist[[12]],objectlist[[13]],objectlist[[14]],objectlist[[15]],objectlist[[16]]),add.cell.ids=ids, merge.data=T)#need to fix the individual names because they are slightly different from batch1
replacements<- data.frame(from= c("SNG-NA18511.variant2", "SNG-NA18858.variant2", "SNG-NA19160.variant2"), to=c("SNG-NA18511", "SNG-NA18858", "SNG-NA19160"))
merged@meta.data<-FindReplace(merged@meta.data, "individual", replacements, from = "from", to= "to", exact=T, vector=F )Only exact matches will be replaced.#run PCA on full dataset pre-alignment
all.genes= rownames(merged)
merged<-FindVariableFeatures(merged,selection.method="vst", nfeatures = 5000)
#have previously used all genes (nfeatures=25000) and clustering by individual rather than batch (based on proportion of cells per cluster) was still observed downstream. Now using 5000 because it is the upper bound of what has been recommended in the literature.
merged<- ScaleData(merged, features = all.genes)Centering and scaling data matrixmerged<-RunPCA(merged, npcs = 100, verbose=F)DimPlot(merged, reduction = "pca", group.by = "Batch")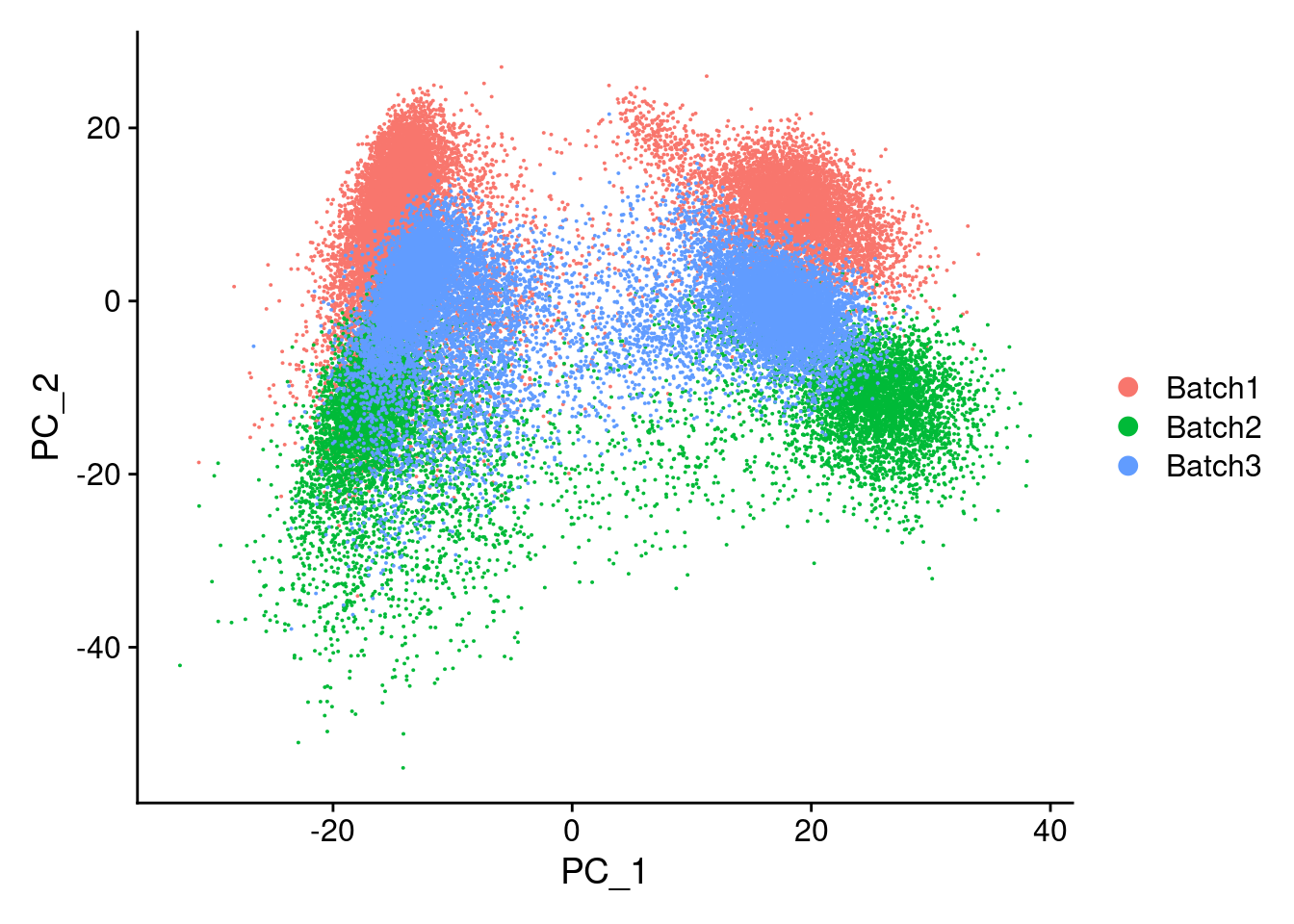
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
Now, running harmony to integrate. Here, using Batch, SampleID(10x Lane), and individual to integrate. Since Batch and Lane are confounded, this may over correct for Batch.
merged<- RunHarmony(merged, c("Batch"), plot_convergence = T, assay.use = "SCT")Harmony 1/10Harmony 2/10Harmony 3/10Harmony 4/10Harmony 5/10Harmony 6/10Harmony 7/10Harmony converged after 7 iterationsWarning: Invalid name supplied, making object name syntactically valid. New
object name is Seurat..ProjectDim.SCT.harmony; see ?make.names for more details
on syntax validity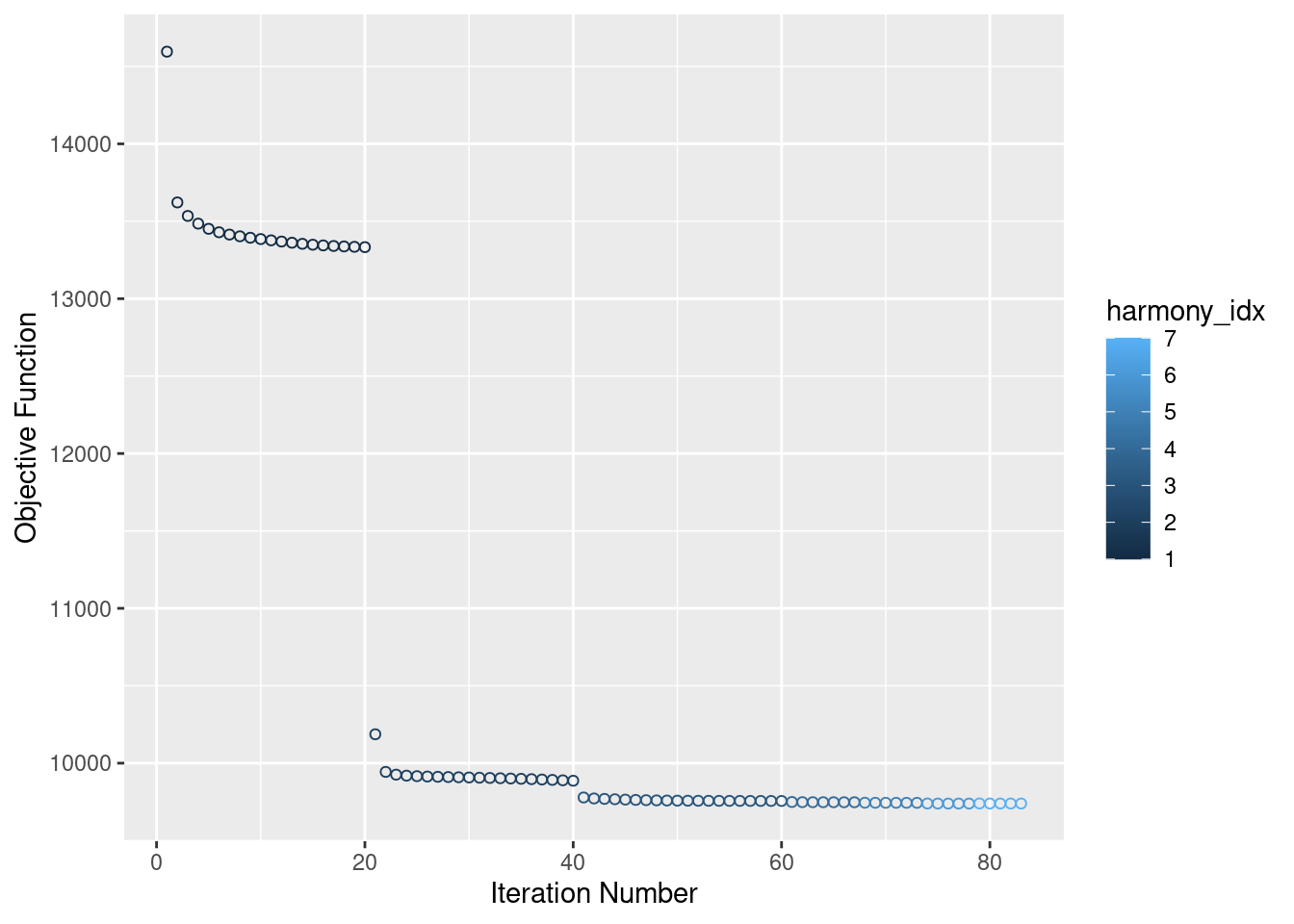
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
Visualize Harmony embeddings
DimPlot(merged, reduction="harmony", group.by= c("individual", "Batch"), combine=F)[[1]]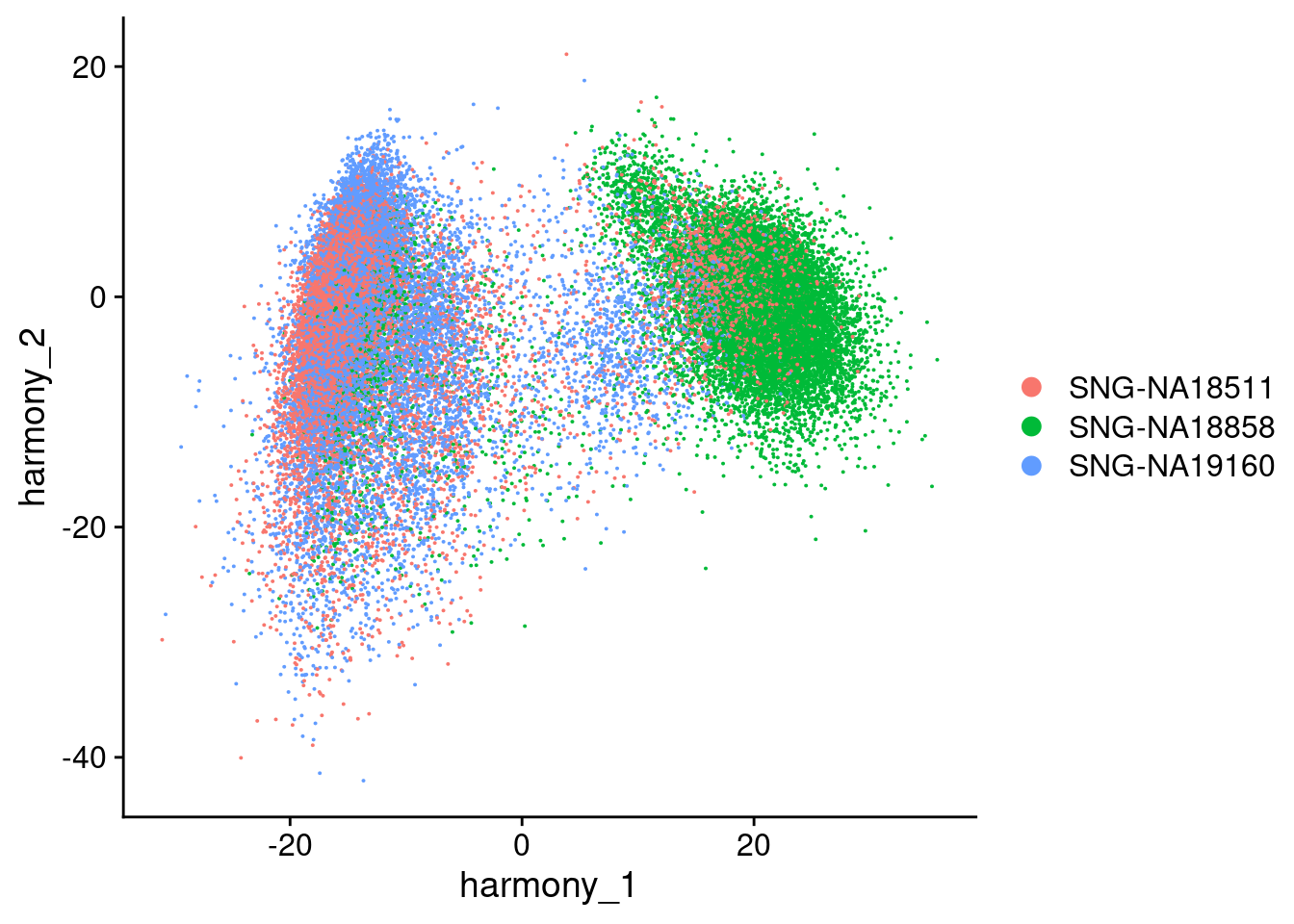
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
[[2]]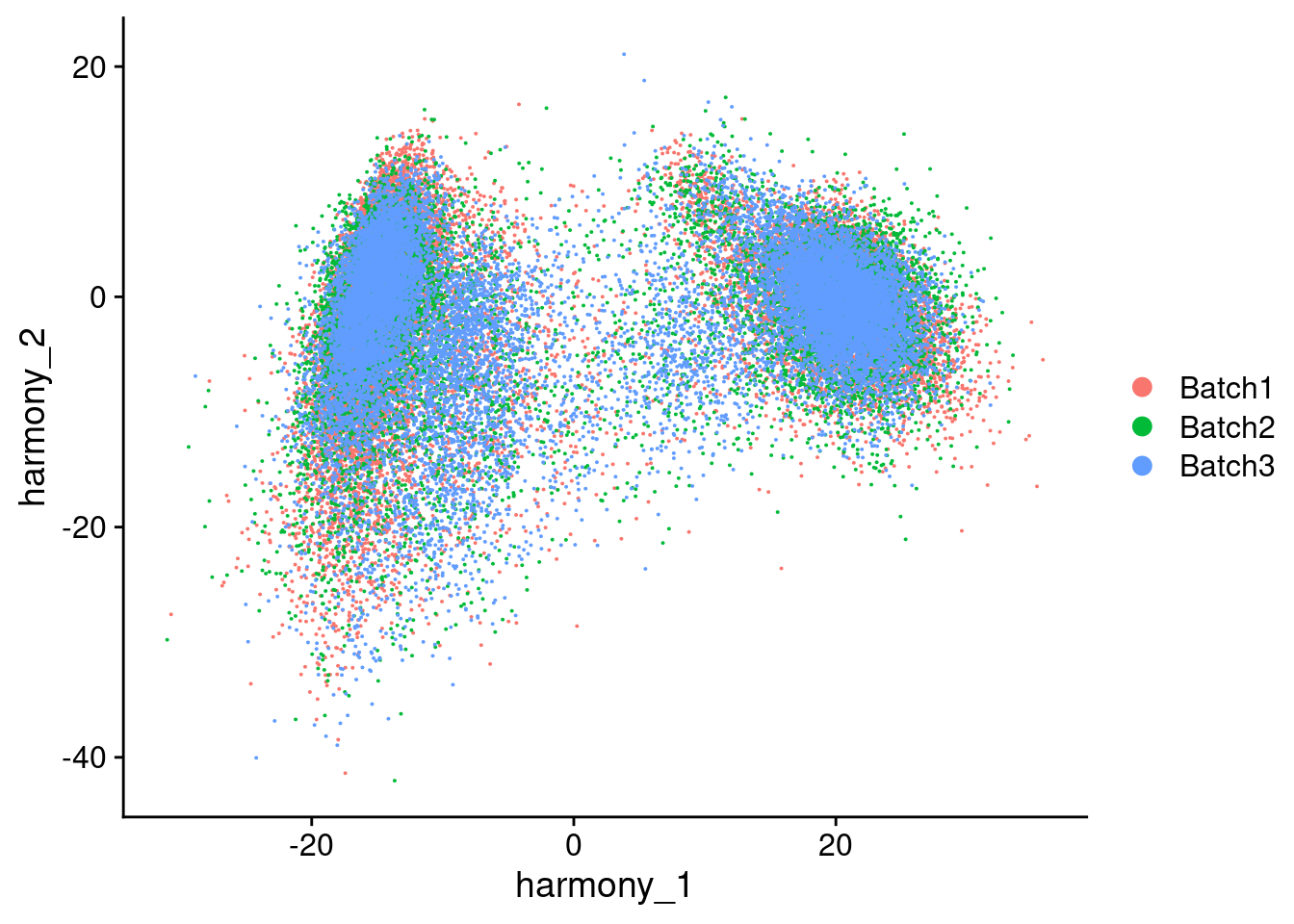
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
Now Running UMAP and identifying clusters, etc
merged<- RunUMAP(merged, reduction = "harmony", dims = 1:100, verbose = F)Warning: The default method for RunUMAP has changed from calling Python UMAP via reticulate to the R-native UWOT using the cosine metric
To use Python UMAP via reticulate, set umap.method to 'umap-learn' and metric to 'correlation'
This message will be shown once per sessionmerged<- FindNeighbors(merged, reduction="harmony", dims = 1:100, verbose = F)
merged<- FindClusters(merged, resolution=1, verbose = F)
merged<- FindClusters(merged, resolution=0.8, verbose = F)
merged<- FindClusters(merged, resolution=0.5, verbose = F)
merged<- FindClusters(merged, resolution=0.1, verbose = F)SAVING merged/aligned/reclustered object
path<- here::here("output/mergedObjects/")
saveRDS(merged, file=paste0(path,'Harmony.Batch.rds'))#reassign idents
Idents(merged)<- 'SCT_snn_res.1'VizDimLoadings(merged, dims = 1:2, reduction = "harmony")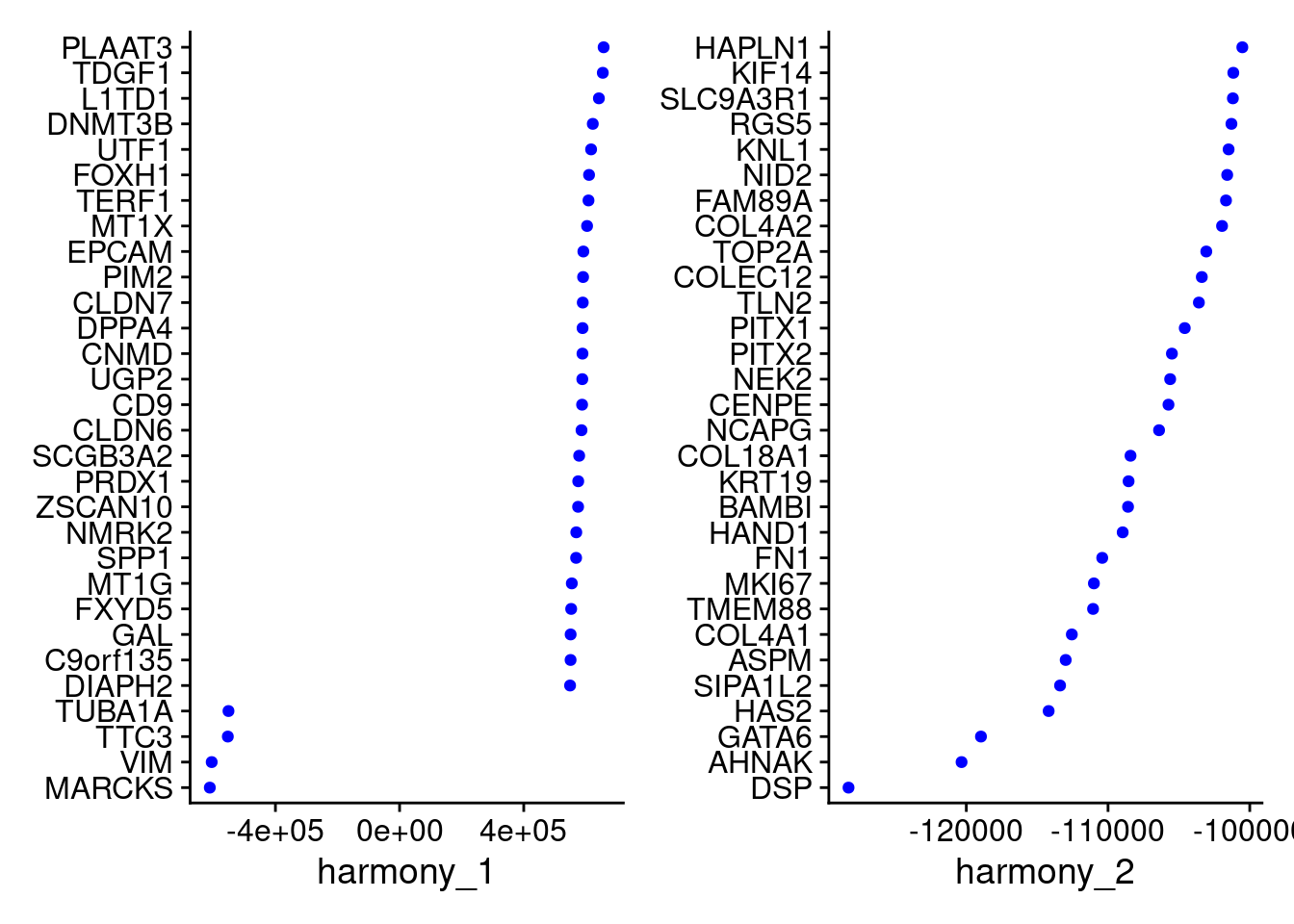
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
VizDimLoadings(merged, dims = 3:4, reduction = "harmony")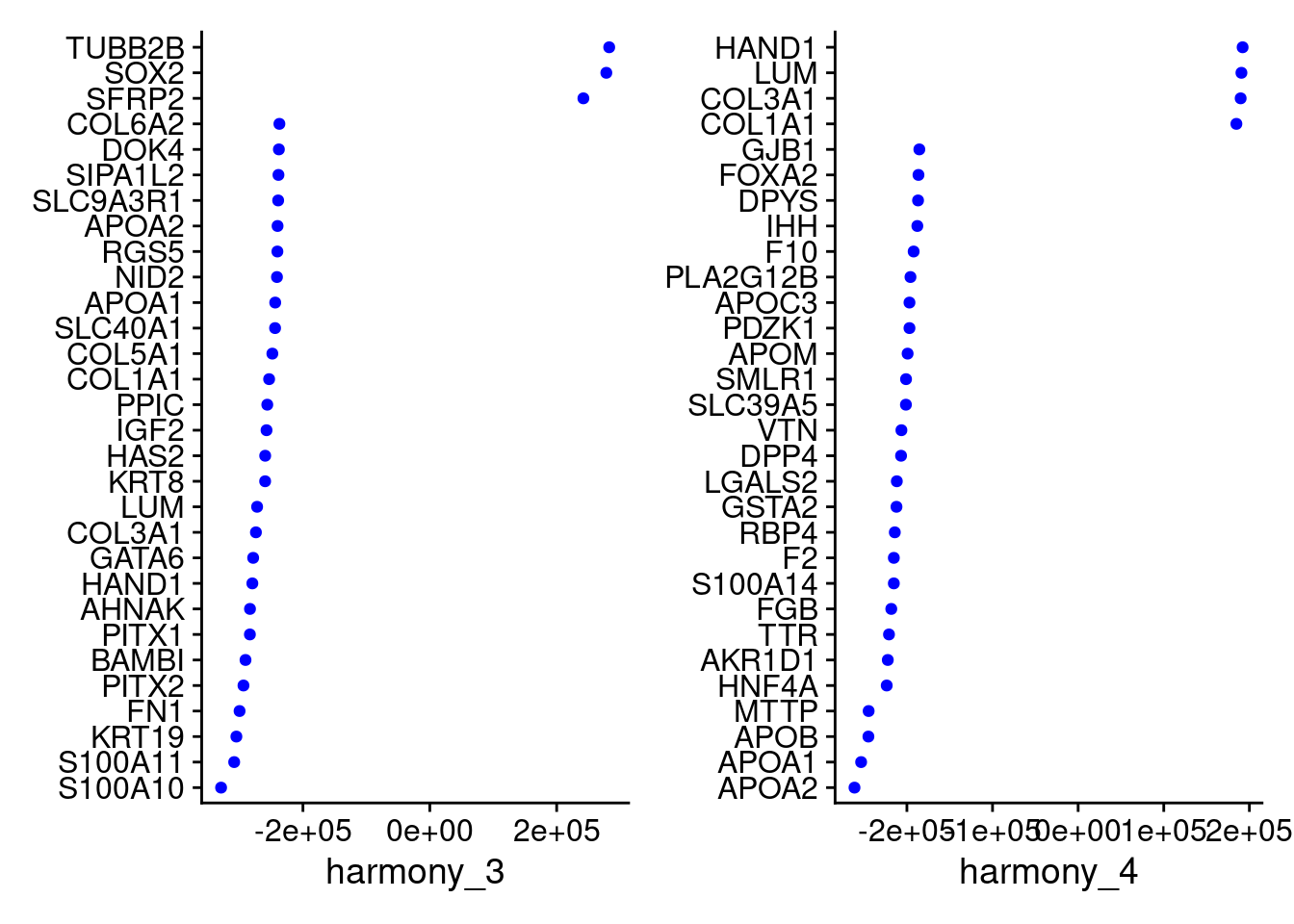
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
VizDimLoadings(merged, dims = 5:6, reduction = "harmony")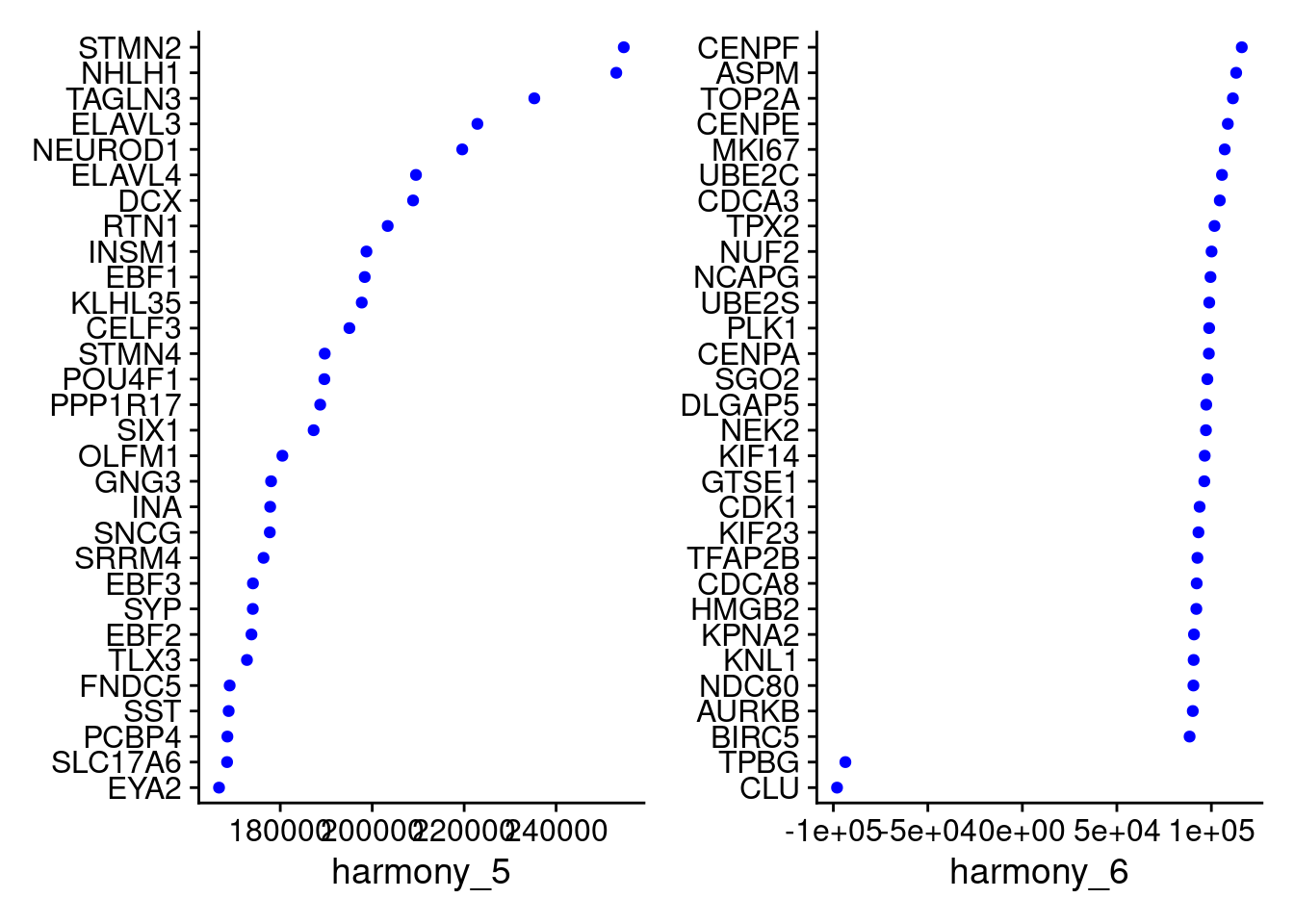
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
xlim <- c(min(merged@reductions$harmony@cell.embeddings[,'harmony_1']),
max(merged@reductions$harmony@cell.embeddings[,'harmony_1']))
ylim <- c(min(merged@reductions$harmony@cell.embeddings[,'harmony_2']),
max(merged@reductions$harmony@cell.embeddings[,'harmony_2']))
individuals <- table(merged$individual)
individuals <- individuals[individuals>50]
individuals <- names(individuals)
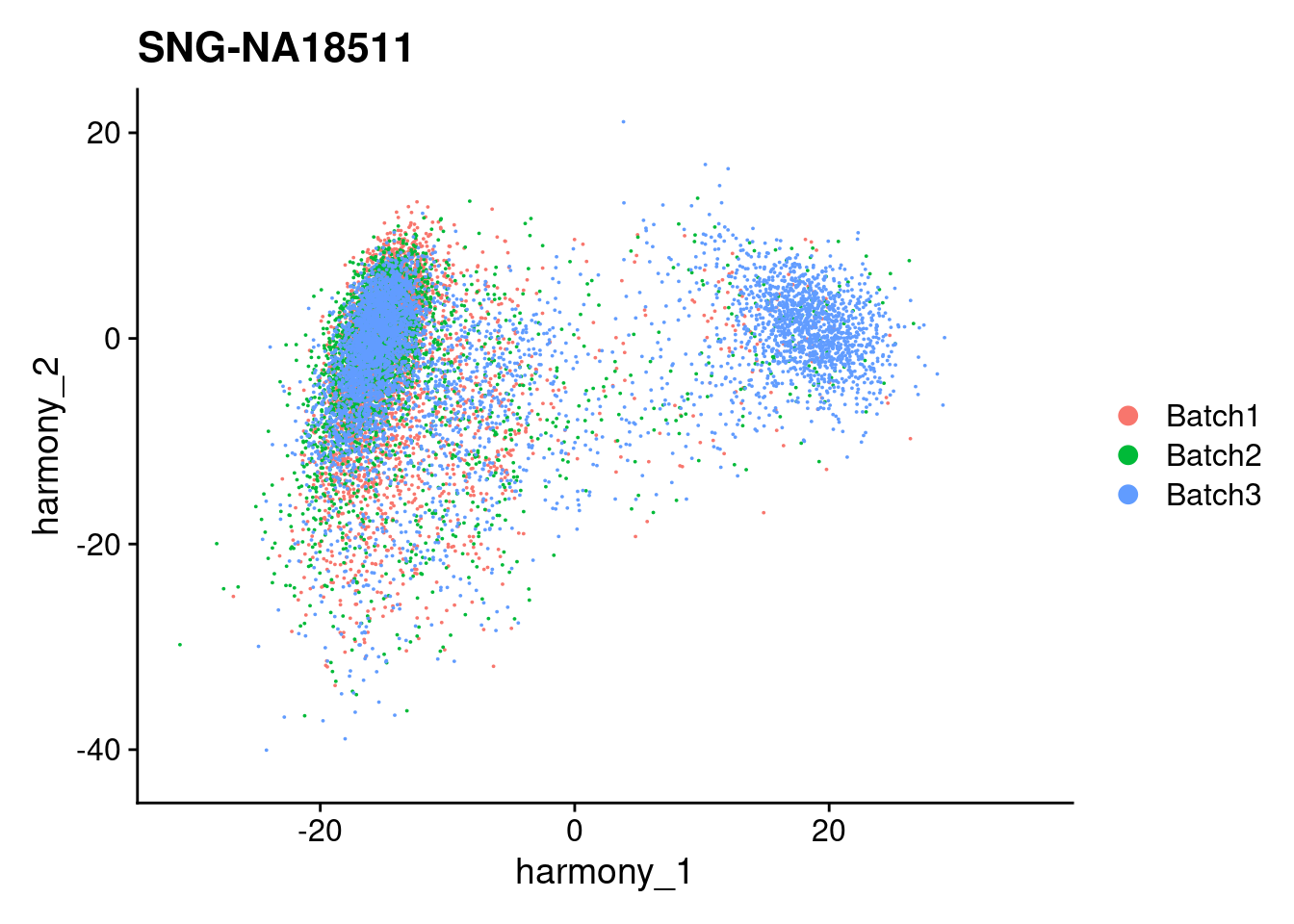
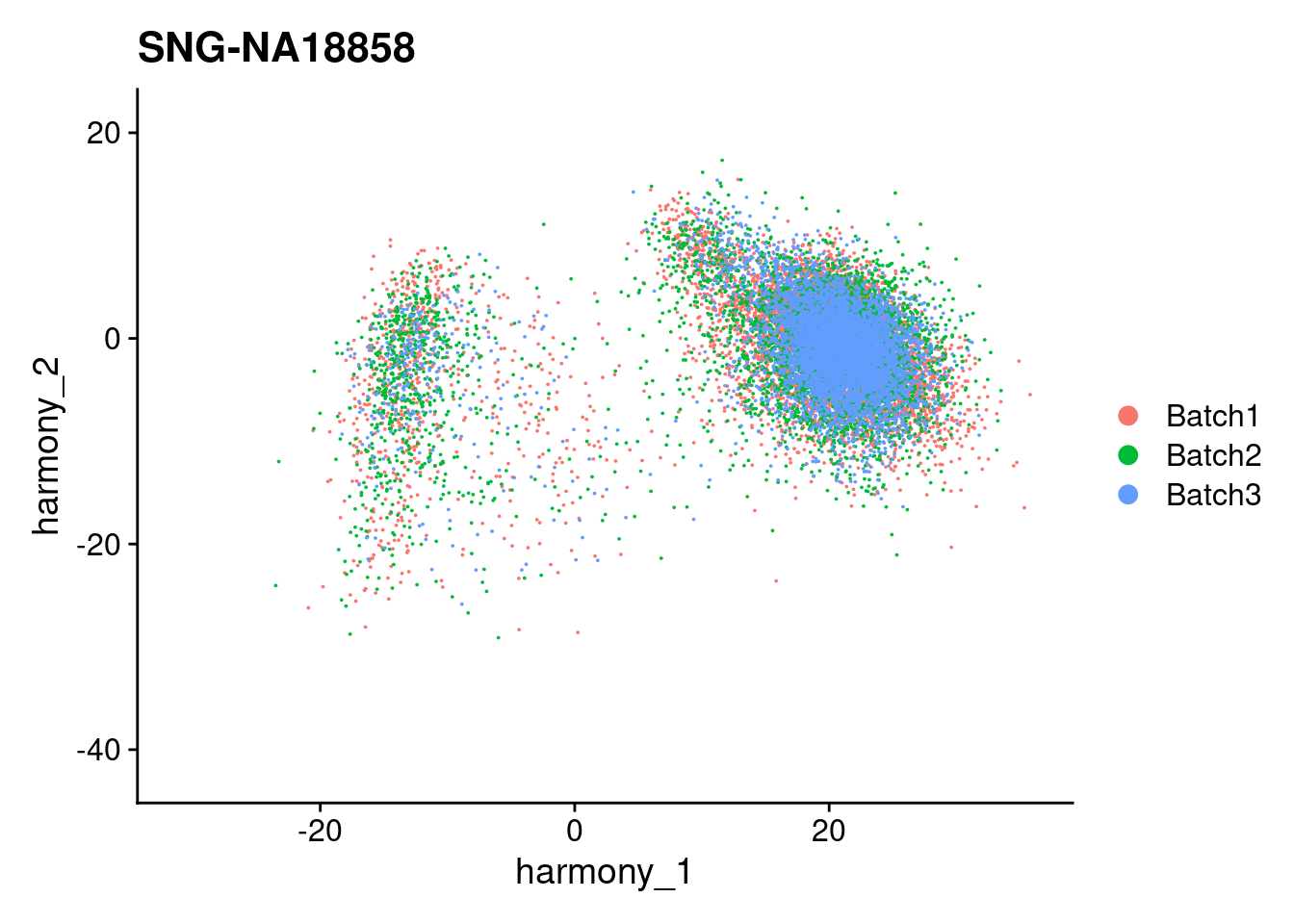
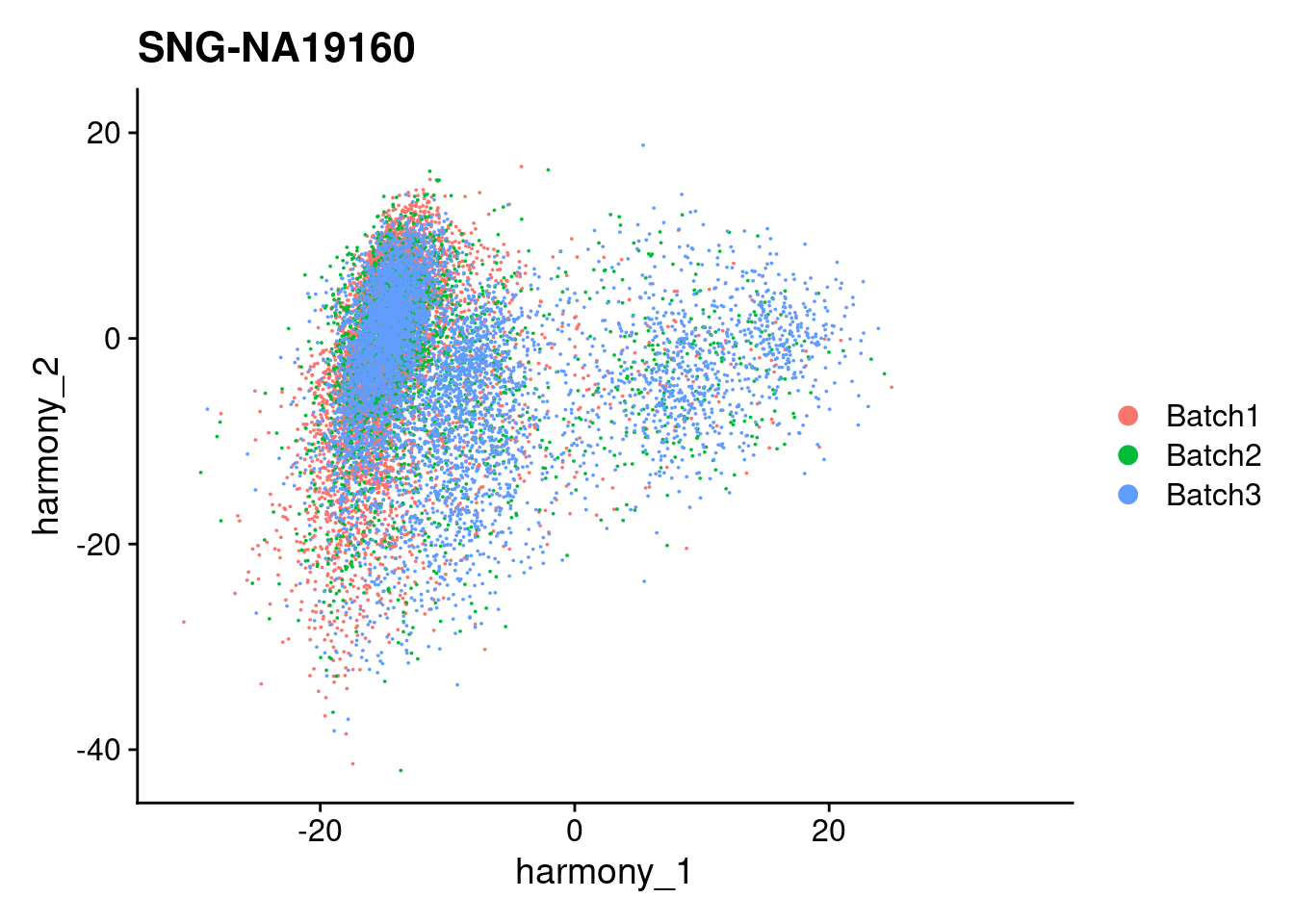
for (i in individuals)
{
print(DimPlot(merged, reduction = "harmony", group.by = c("Batch"), pt.size = 0.01,
cells = WhichCells(merged, expression = individual == i)) +
xlim(xlim) + ylim(ylim) + ggtitle(i))
}
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
DimPlot(merged, reduction = "umap")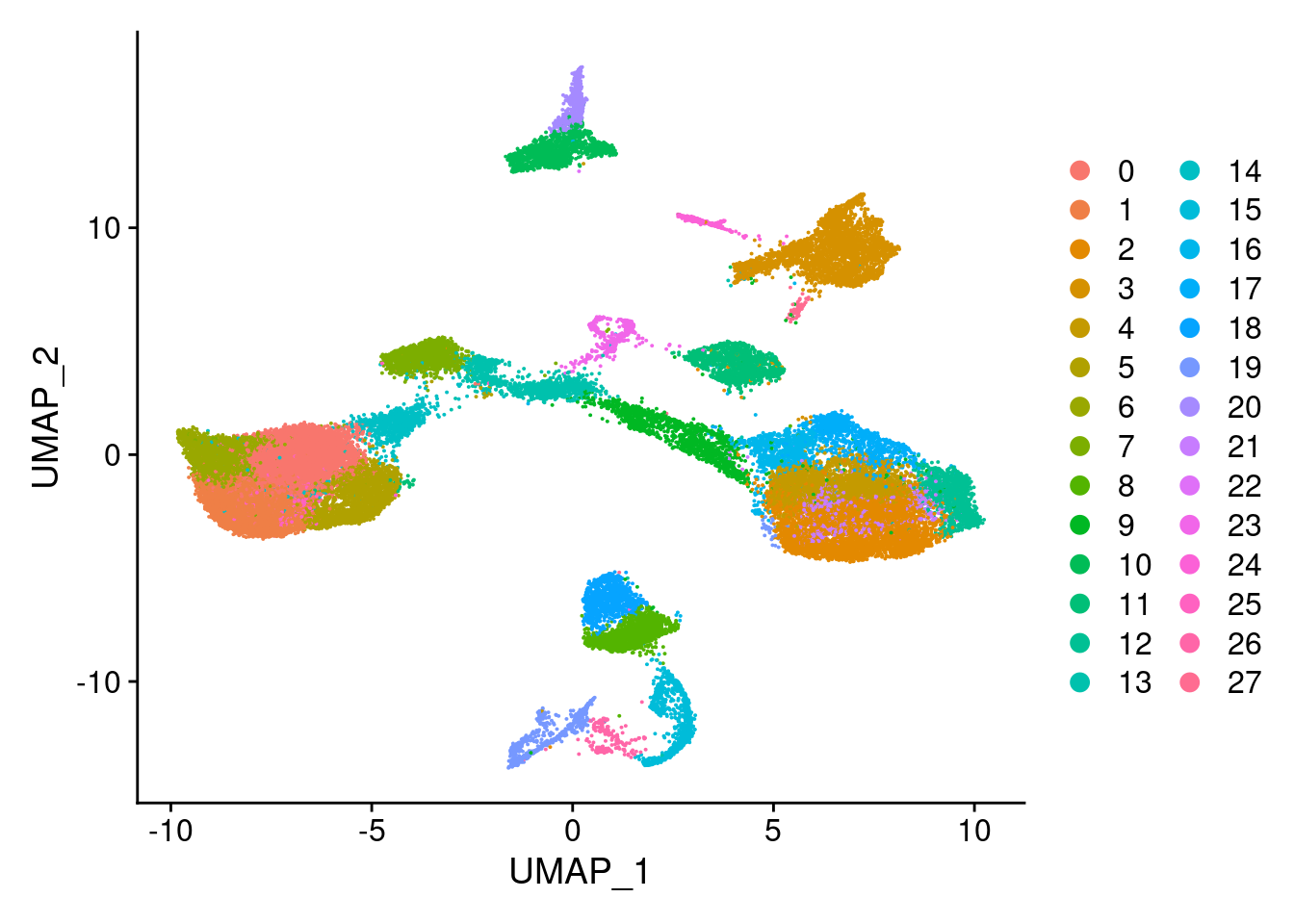
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
DimPlot(merged, reduction = "umap", group.by = "Batch")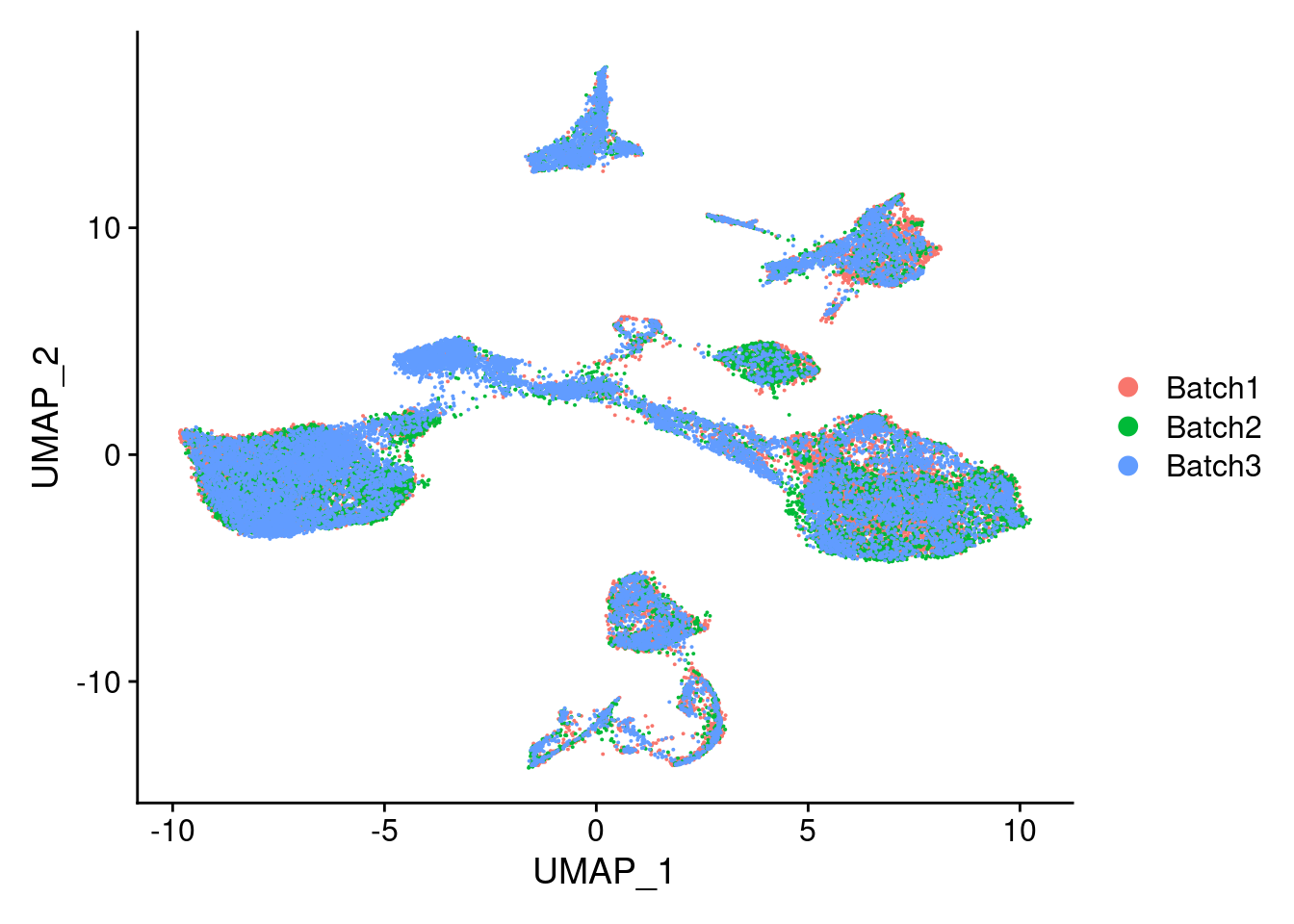
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
DimPlot(merged, reduction = "umap", group.by = "individual")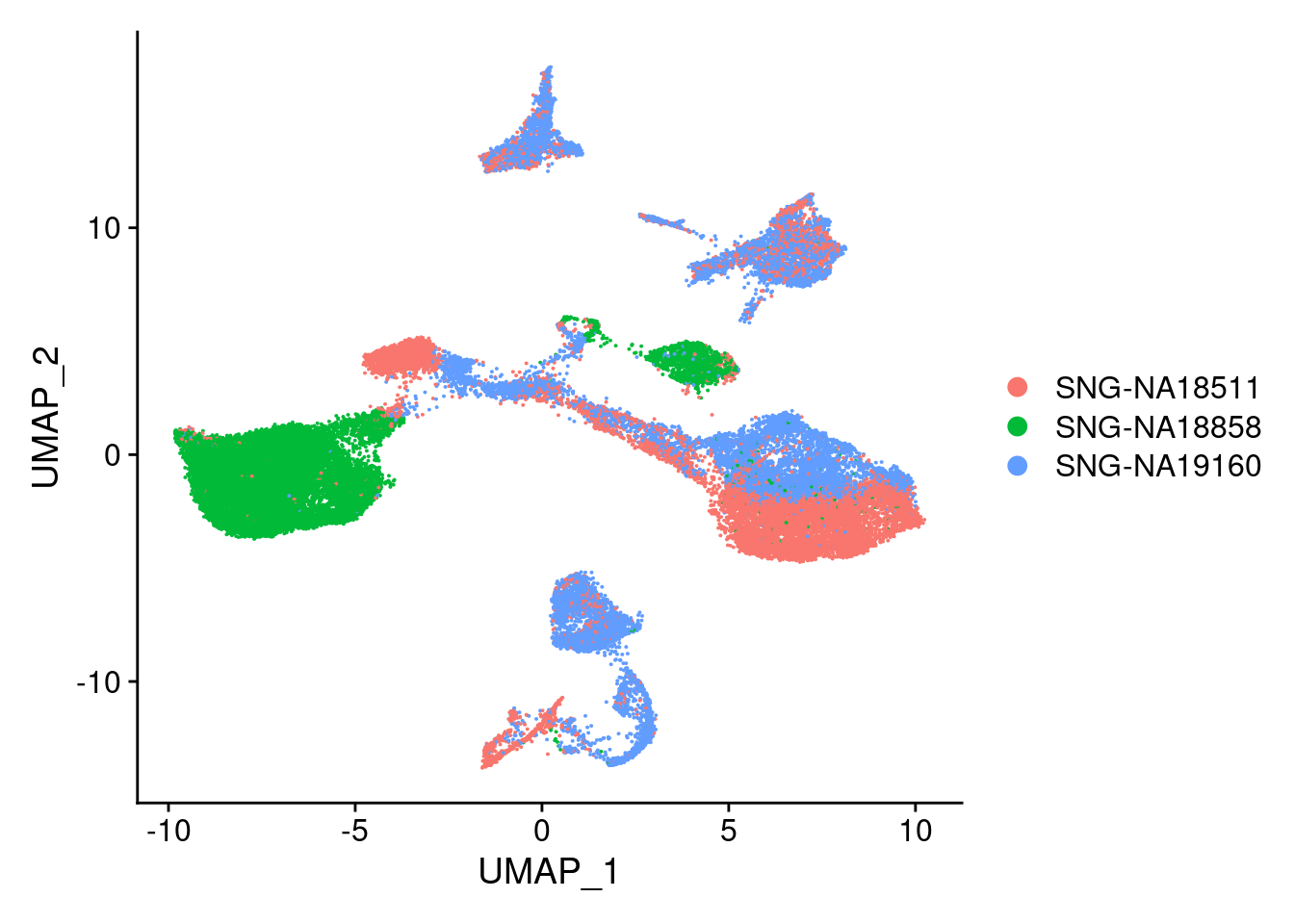
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
xlim <- c(min(merged@reductions$umap@cell.embeddings[,'UMAP_1']),
max(merged@reductions$umap@cell.embeddings[,'UMAP_1']))
ylim <- c(min(merged@reductions$umap@cell.embeddings[,'UMAP_2']),
max(merged@reductions$umap@cell.embeddings[,'UMAP_2']))
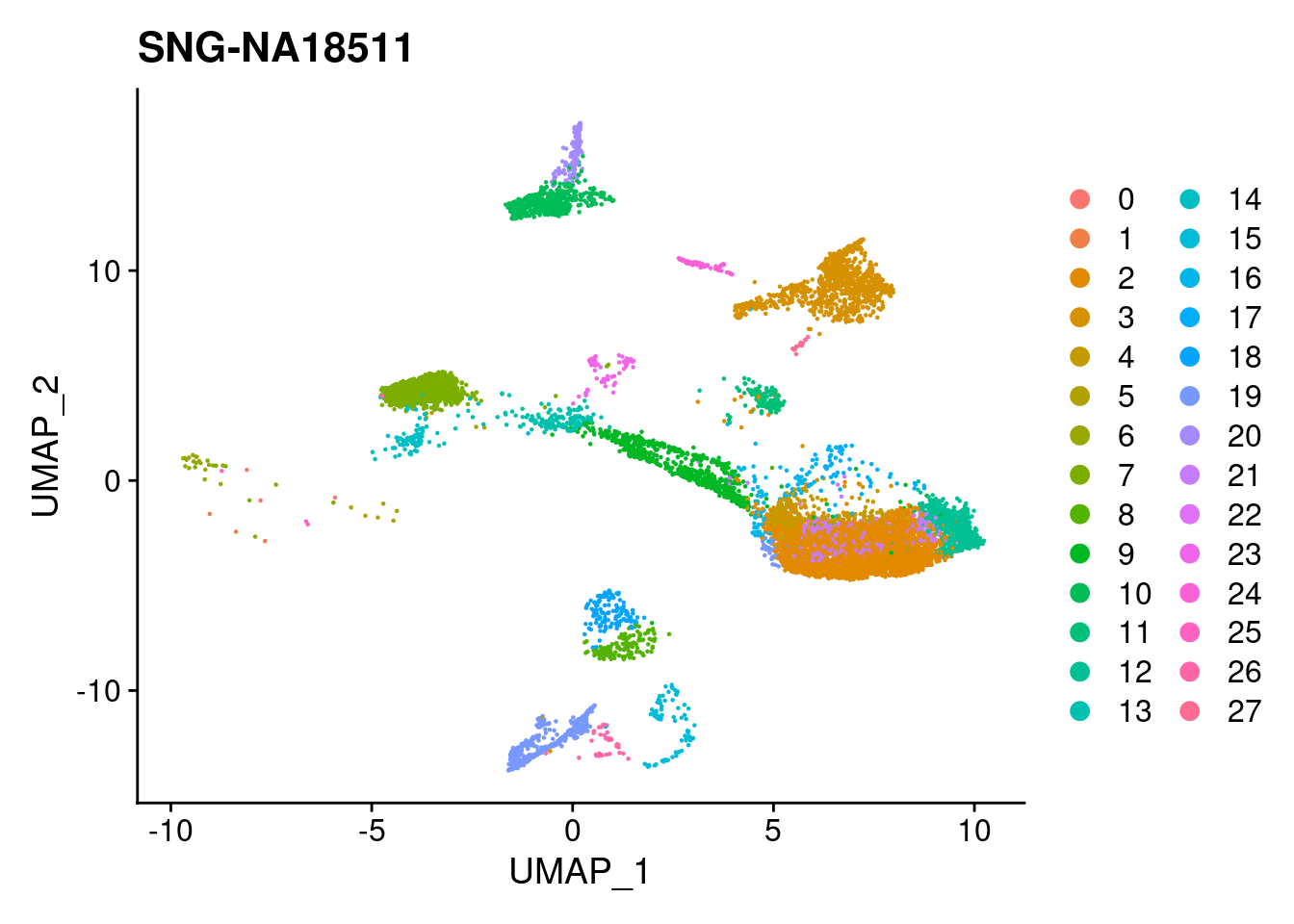
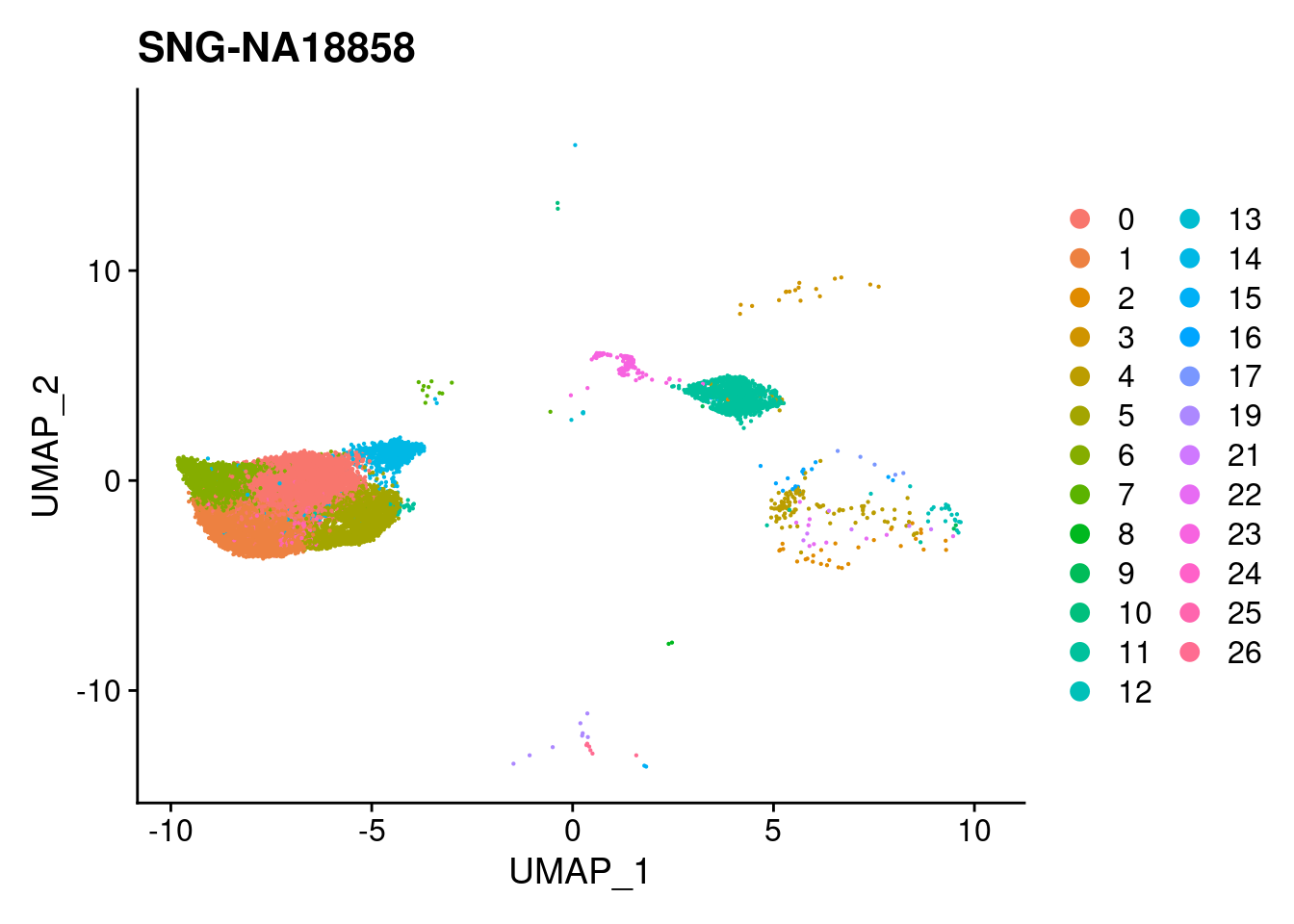
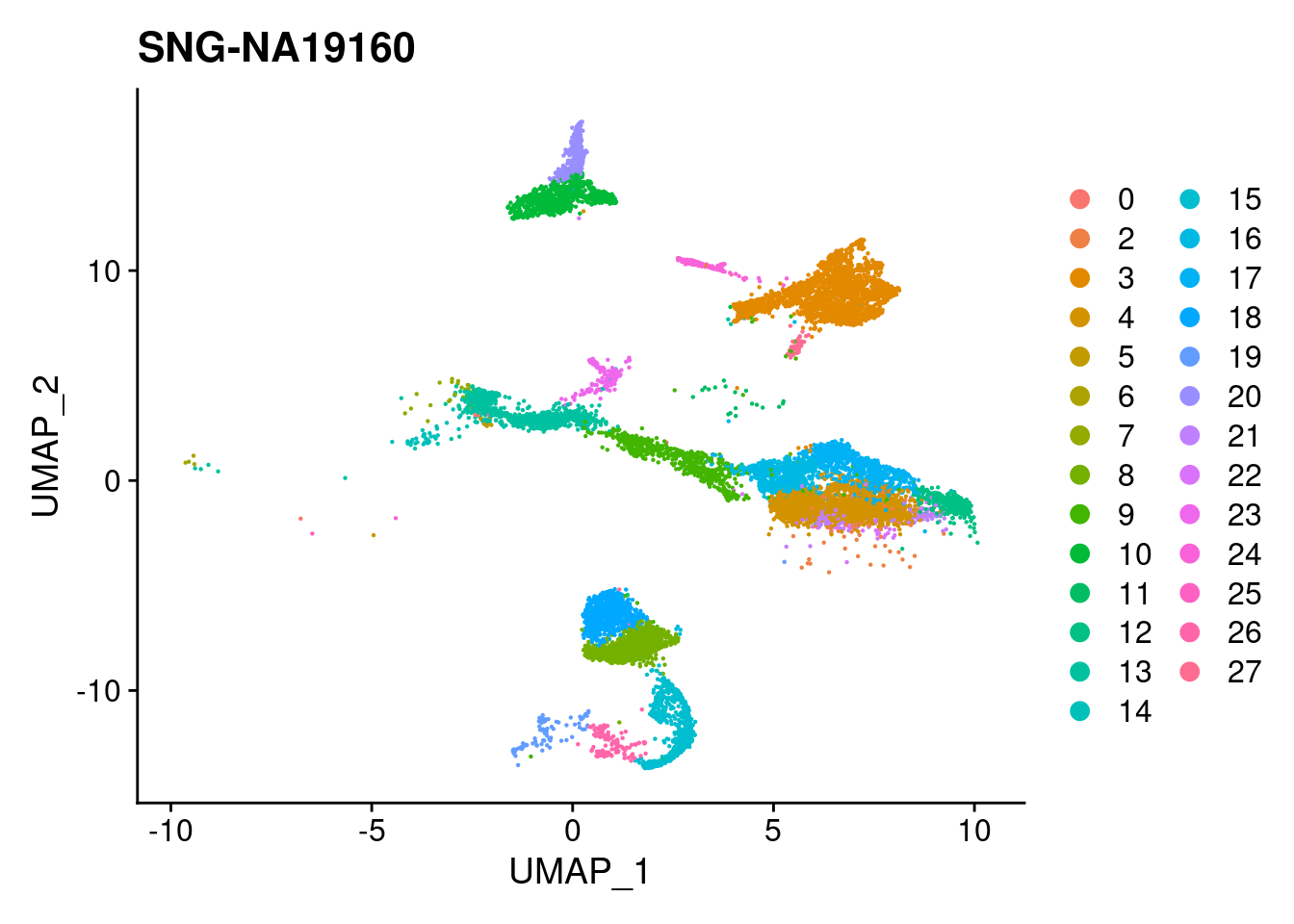
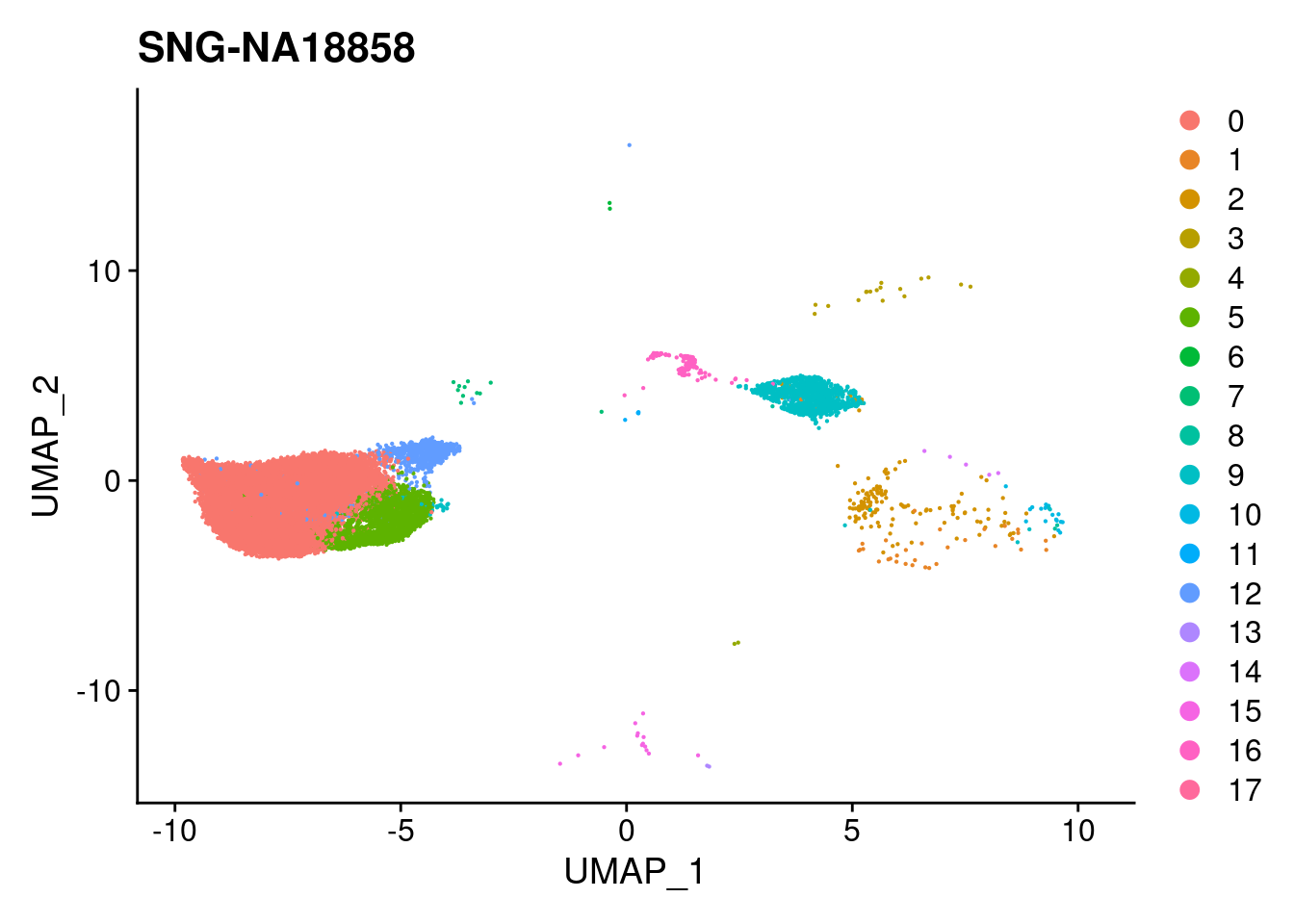
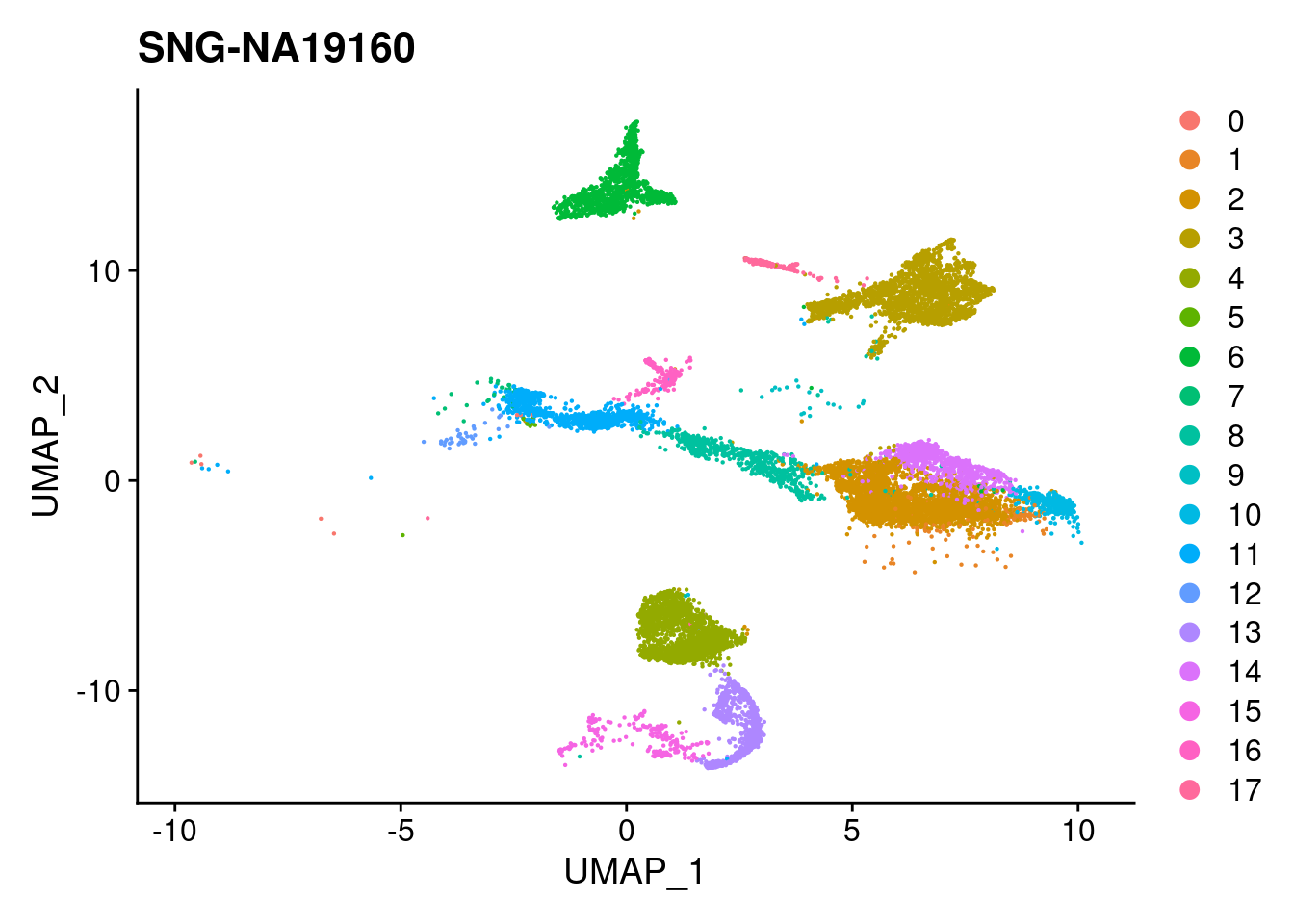
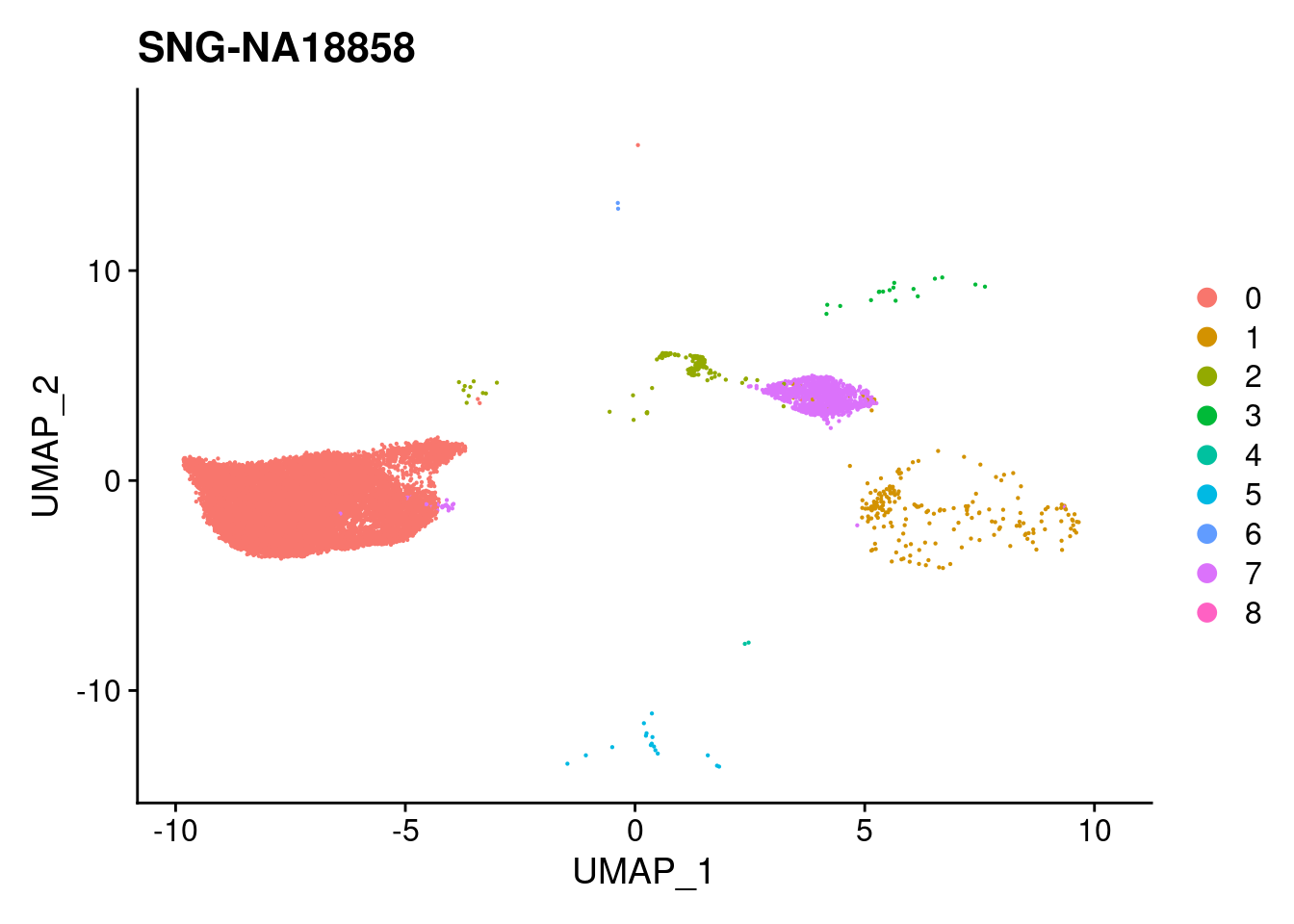
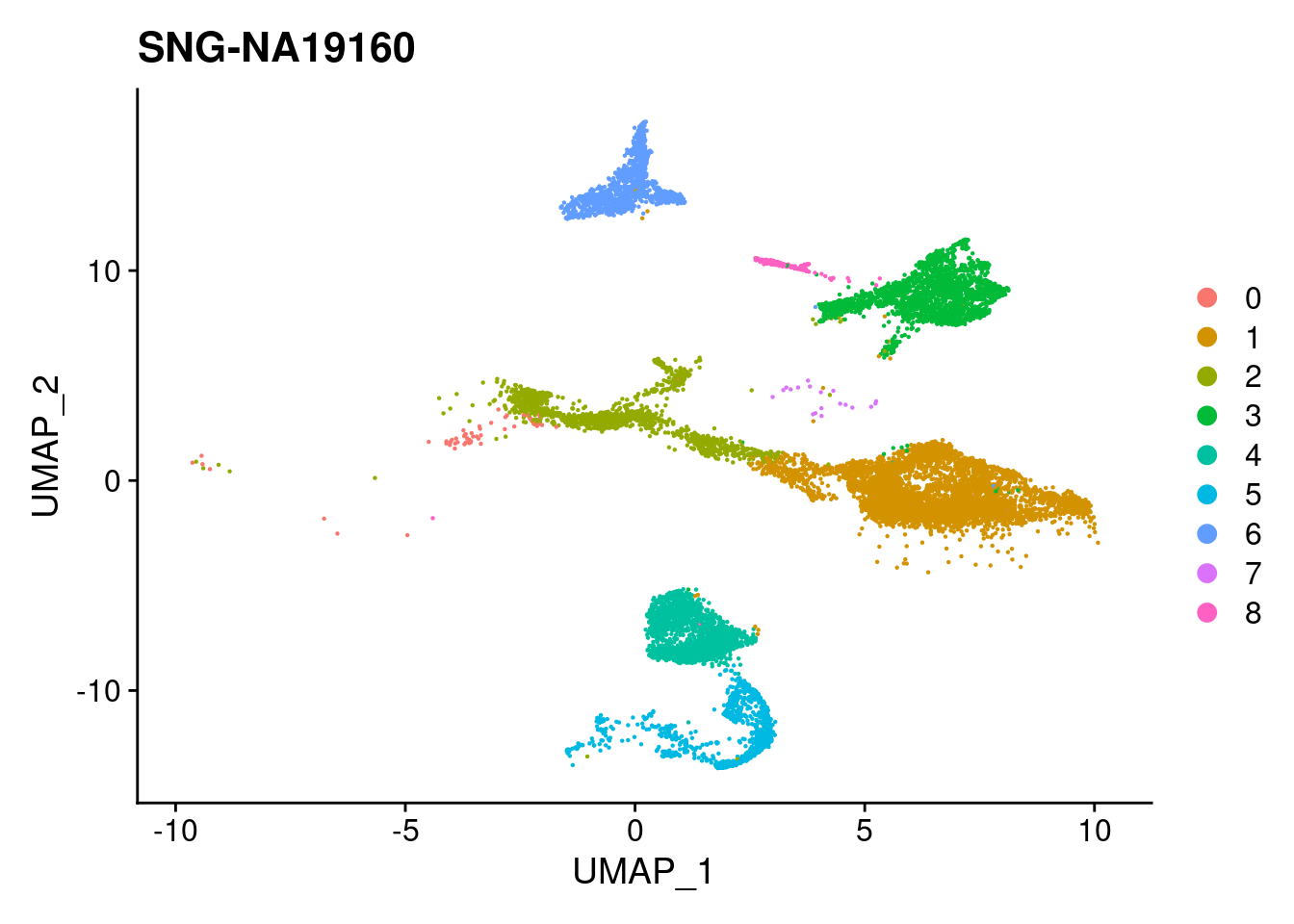
for (i in individuals)
{
print(DimPlot(merged, reduction = "umap",
cells = WhichCells(merged, expression = individual == i)) +
xlim(xlim) + ylim(ylim) + ggtitle(i))
}
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
plots2<- DimPlot(merged, group.by = "individual", split.by = "Batch")
plots2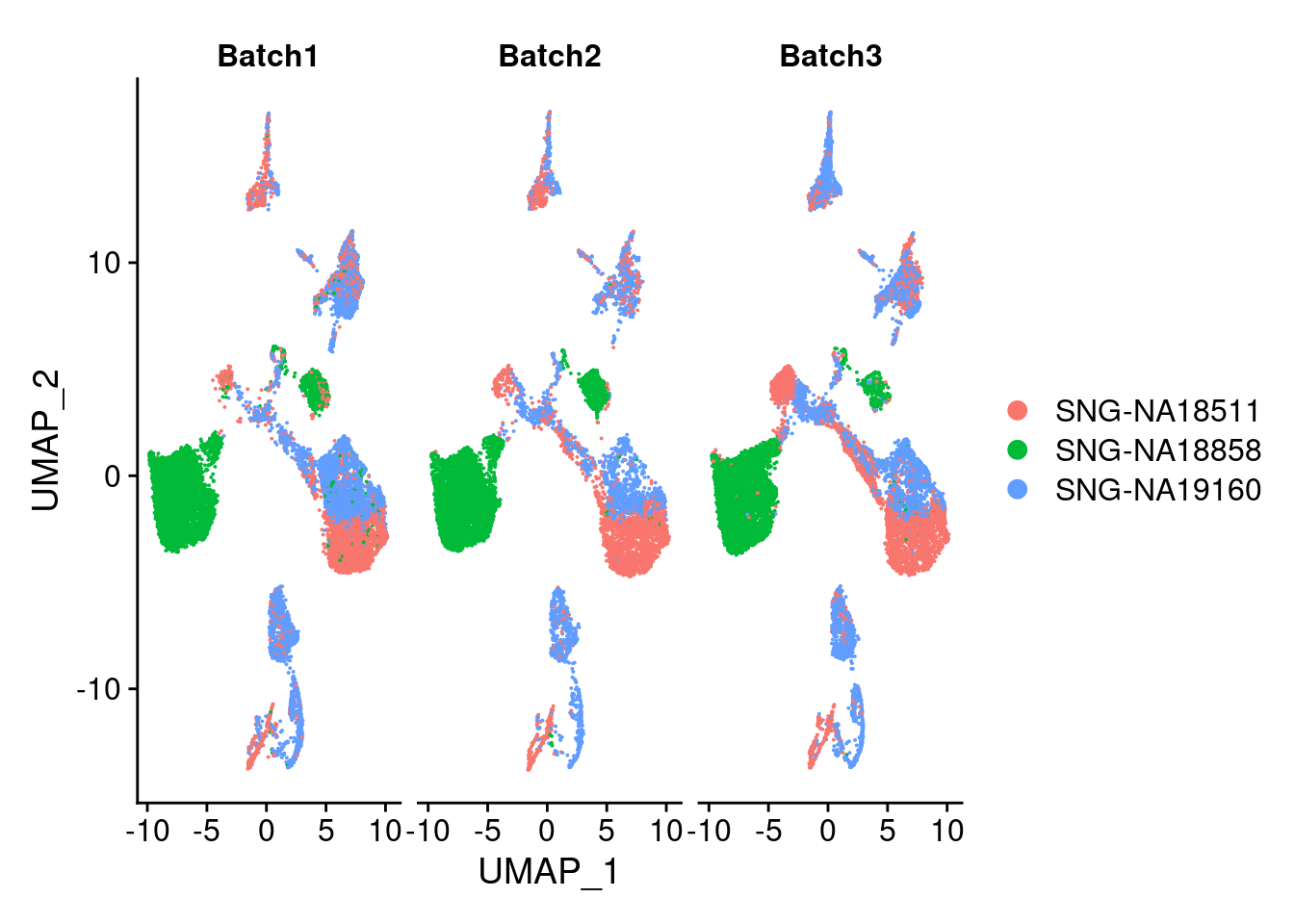
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
DimPlot(merged, group.by = "Batch", split.by = c("individual"))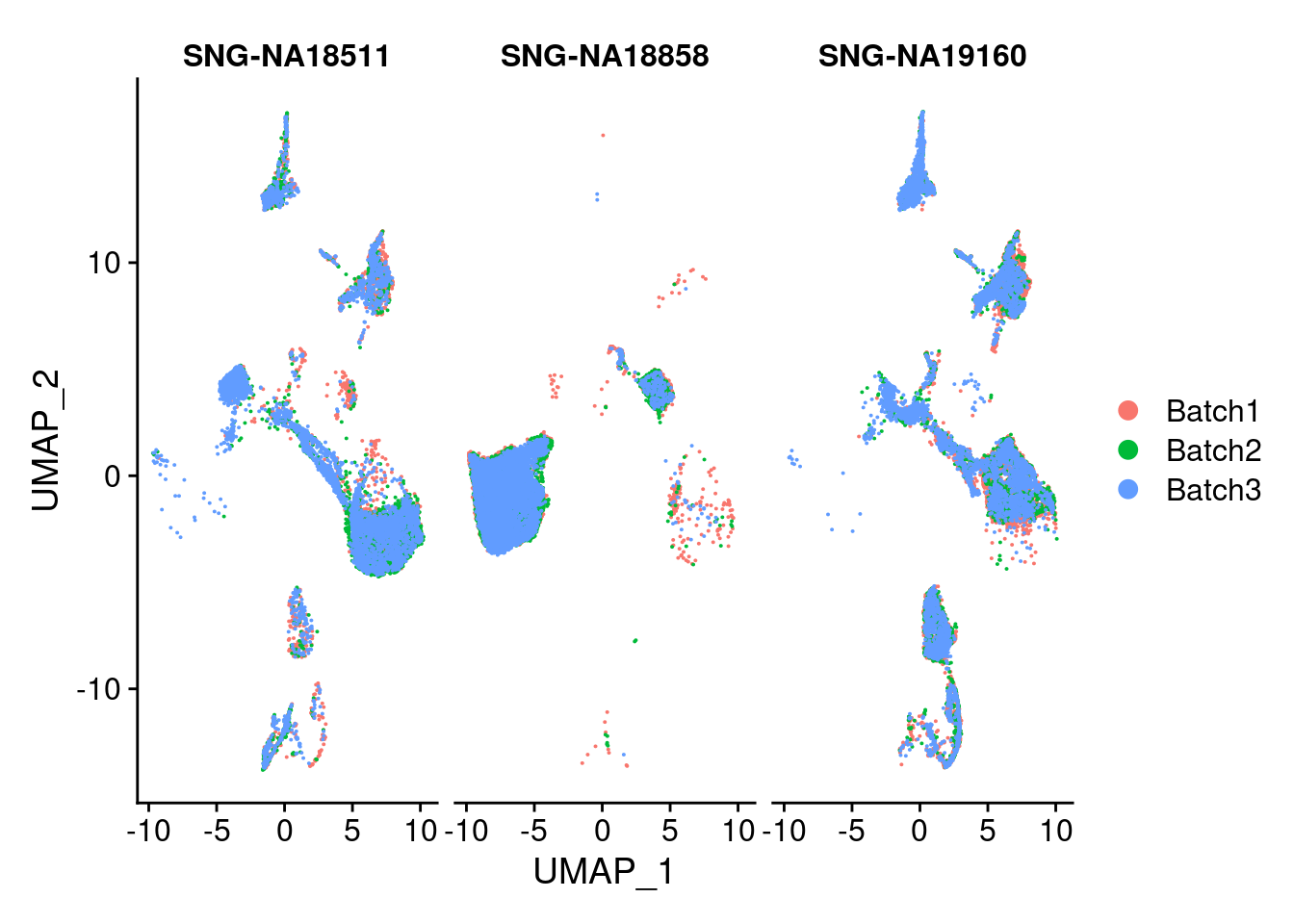
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
DimPlot(merged, group.by = "SCT_snn_res.1", split.by = c("Batch"), label=T)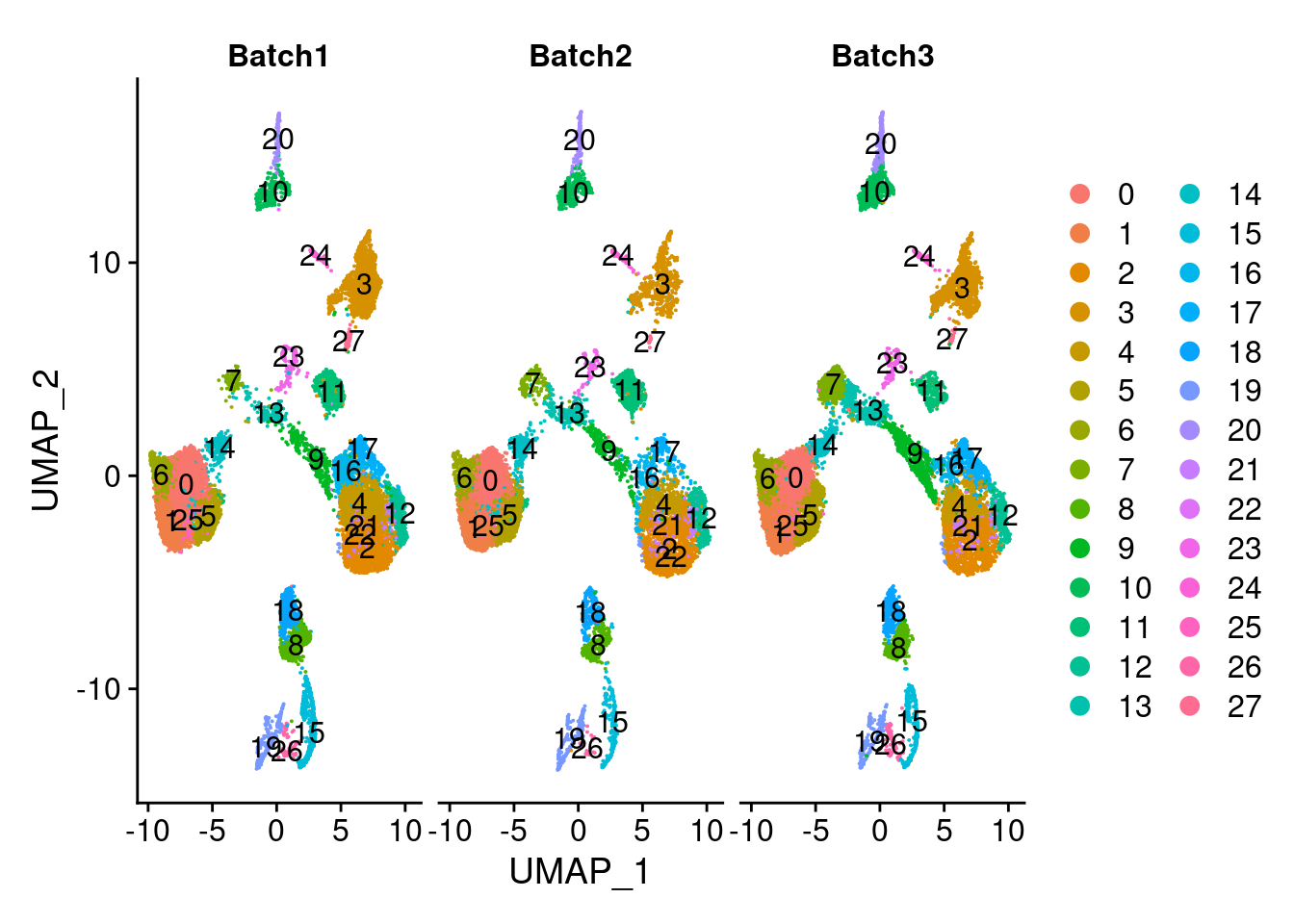
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
DimPlot(merged, reduction = "harmony", group.by = "SCT_snn_res.1", split.by = "Batch", combine = F)[[1]]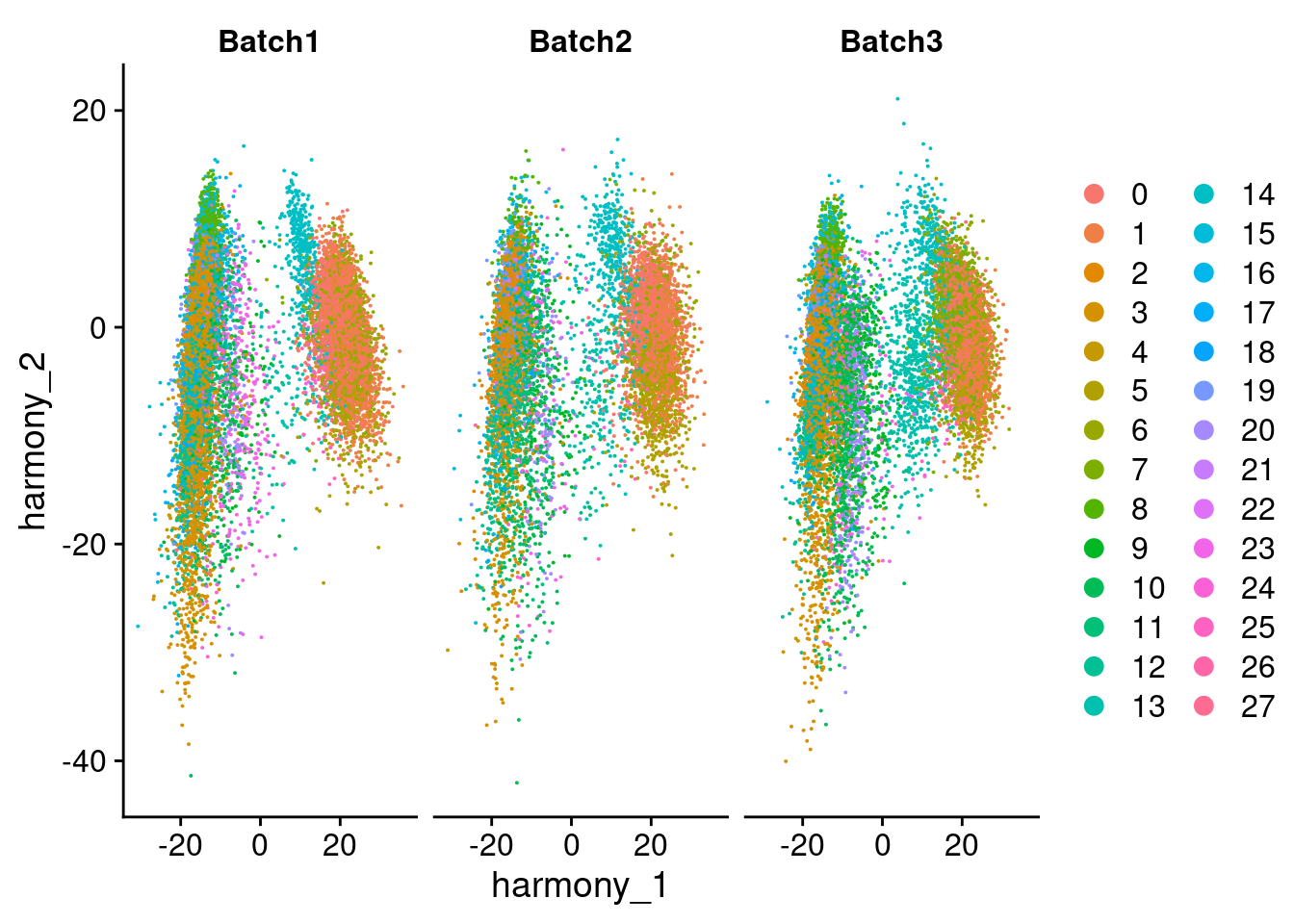
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
VlnPlot(merged, features = c("POU5F1", "PAX6", "TNNT2", "SOX17", "HAND1", "LUM"), ncol=2)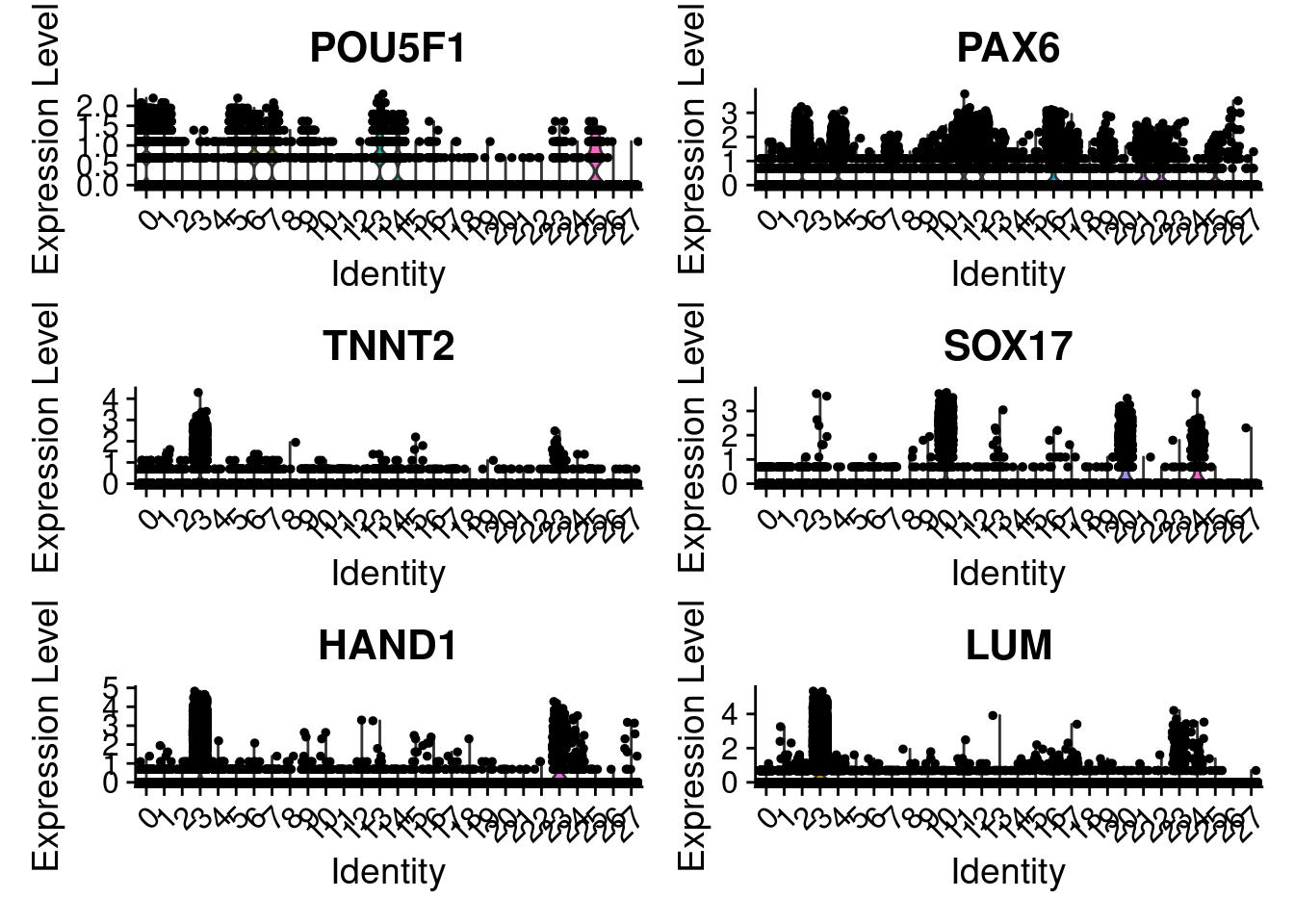
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
#pluripotent markers
FeaturePlot(merged, features = c("POU5F1", "SOX2", "NANOG"), pt.size = 0.2, ncol=3)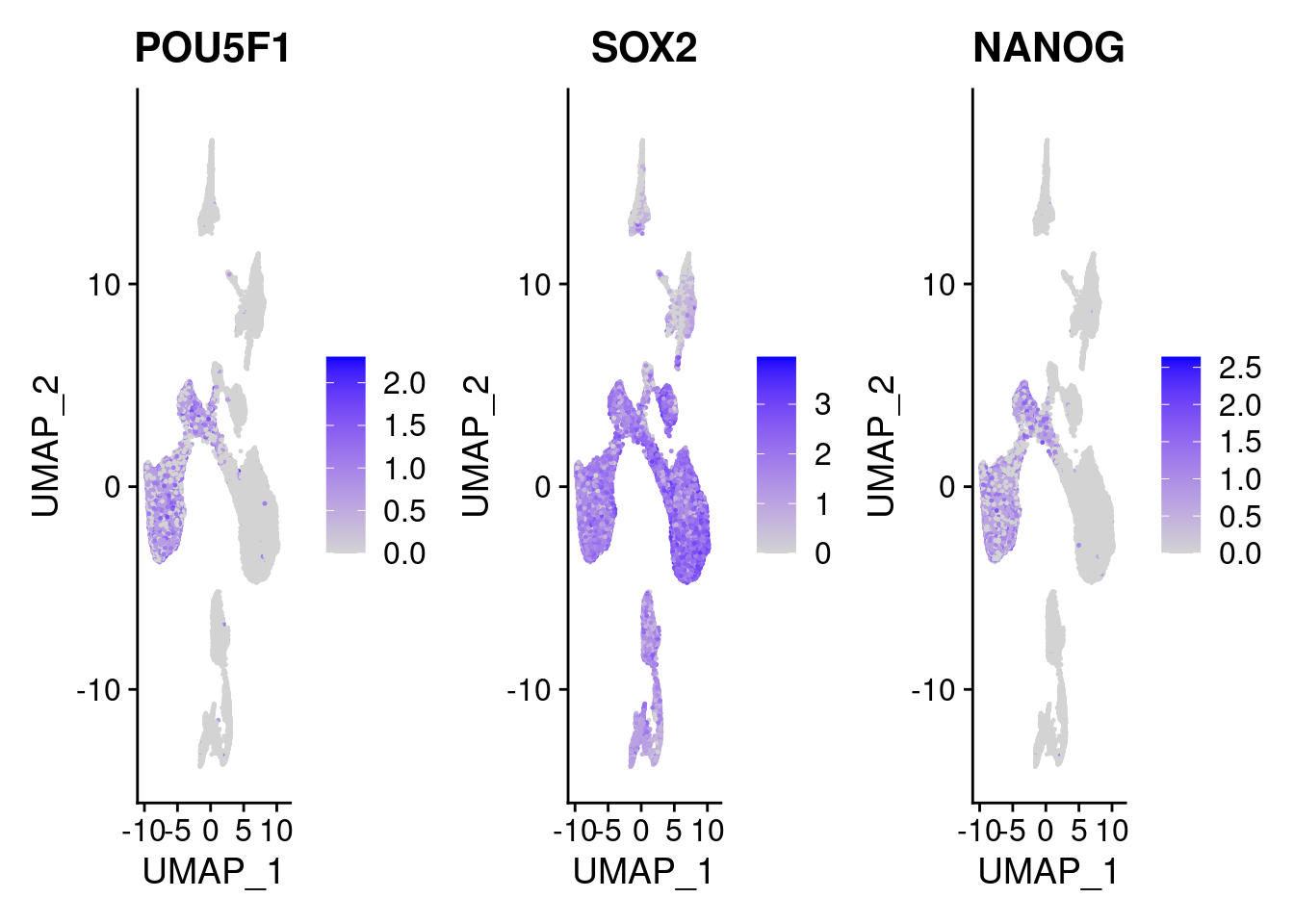
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
#Endoderm markers (first 3 definitive endo, 4-6 liver markers, )
FeaturePlot(merged, features = c("SOX17","CLDN6","FOXA2", "TTR", "AFP", "FGB"), pt.size = 0.2, combine = F)[[1]]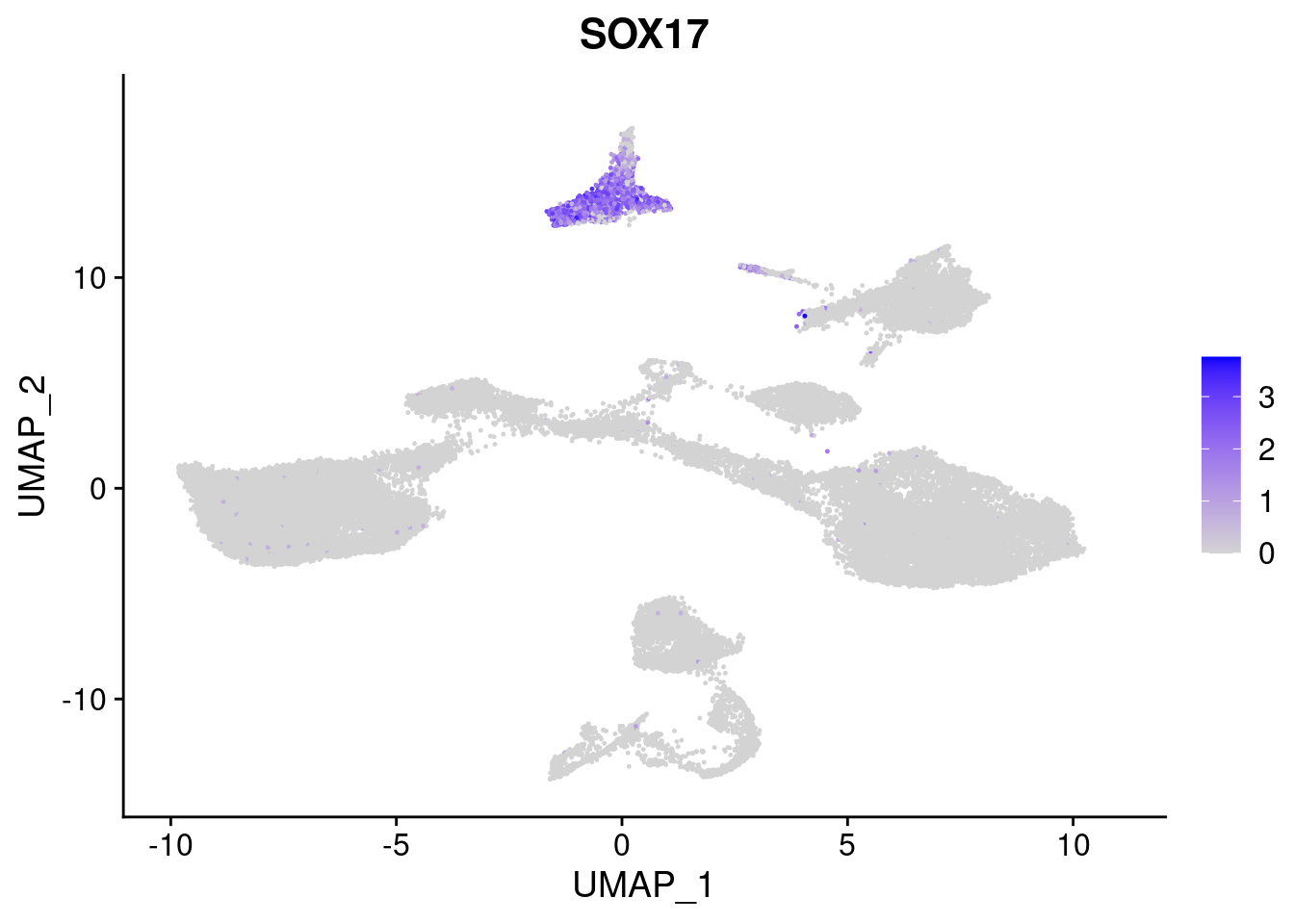
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
[[2]]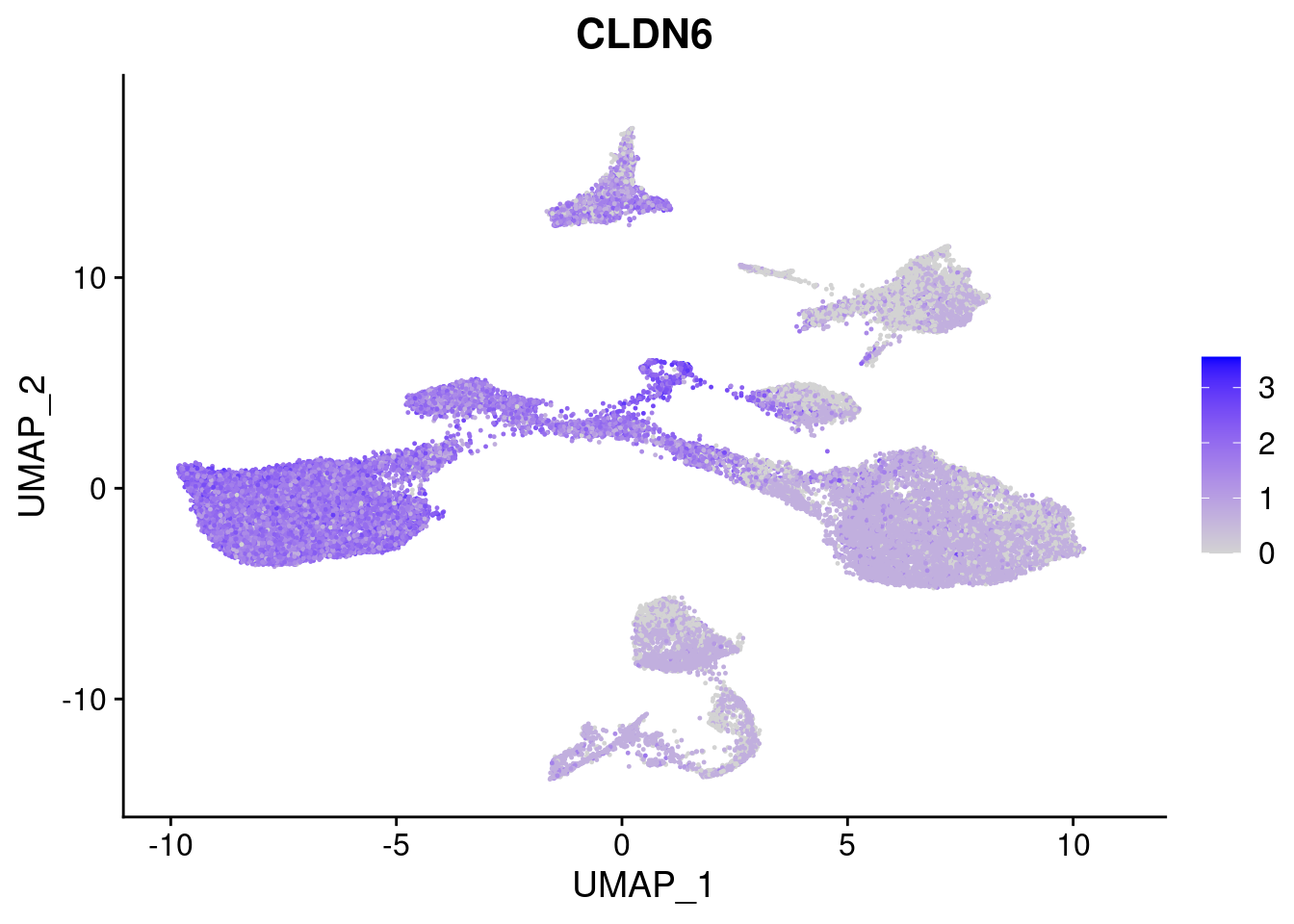
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
[[3]]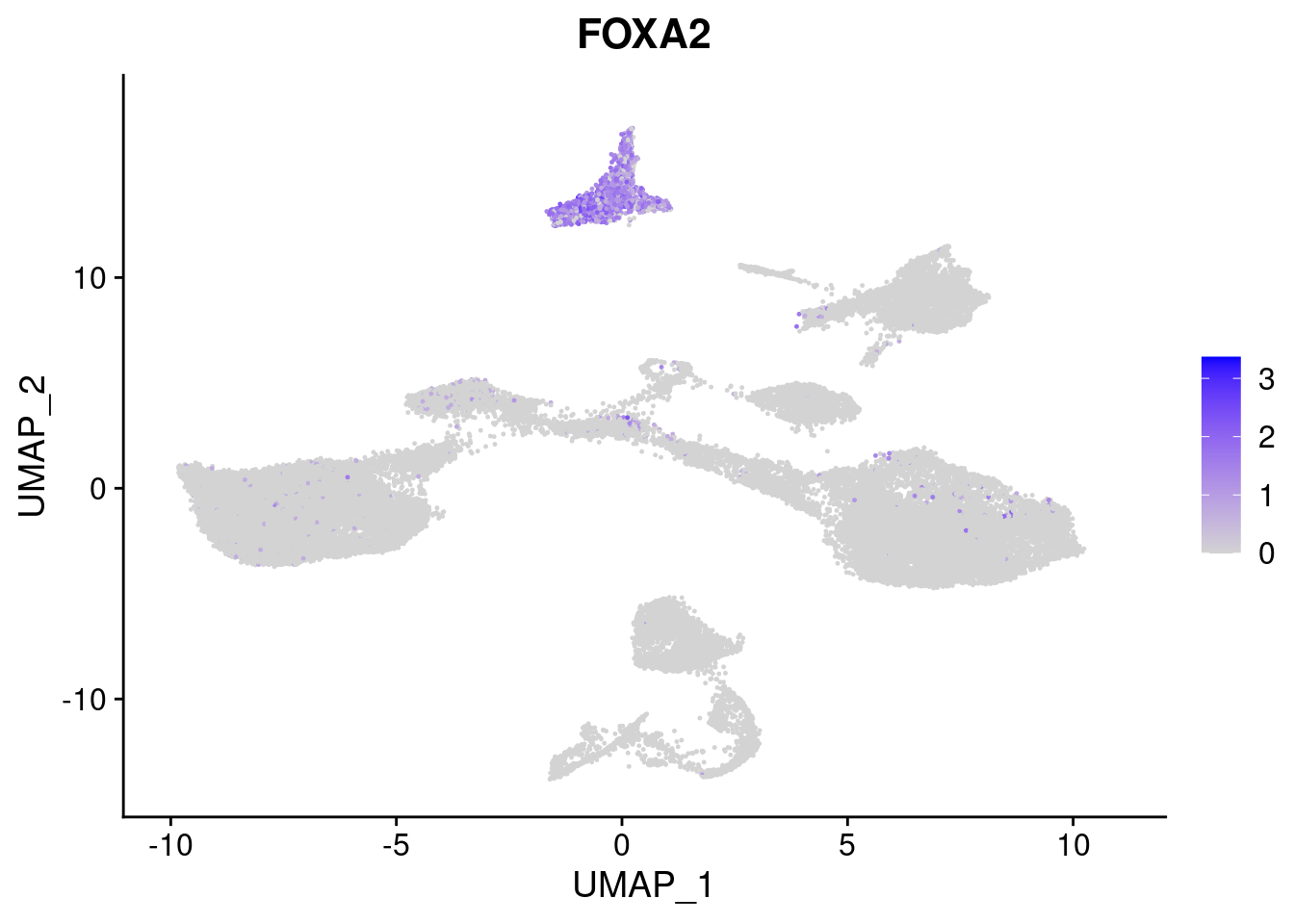
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
[[4]]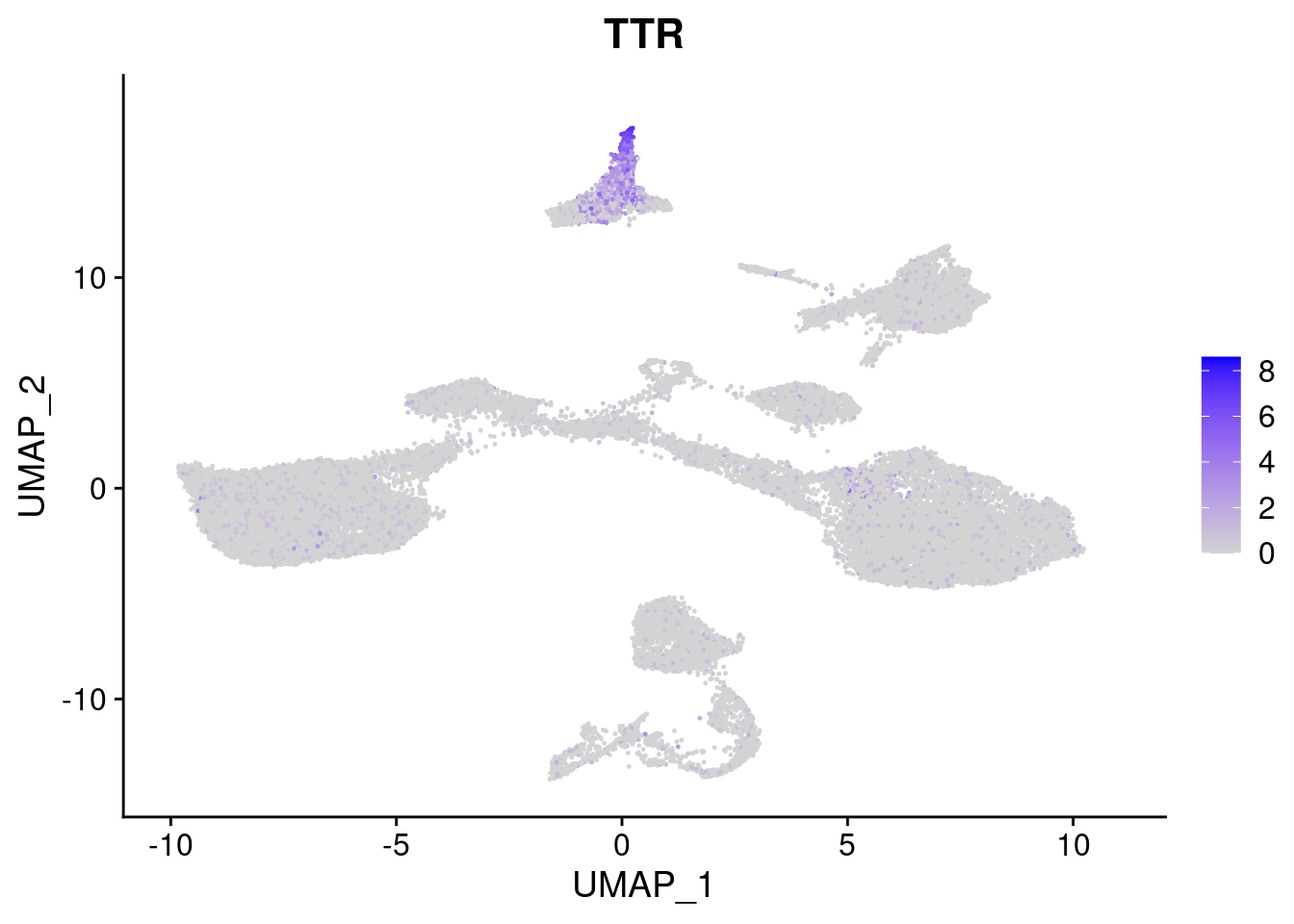
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
[[5]]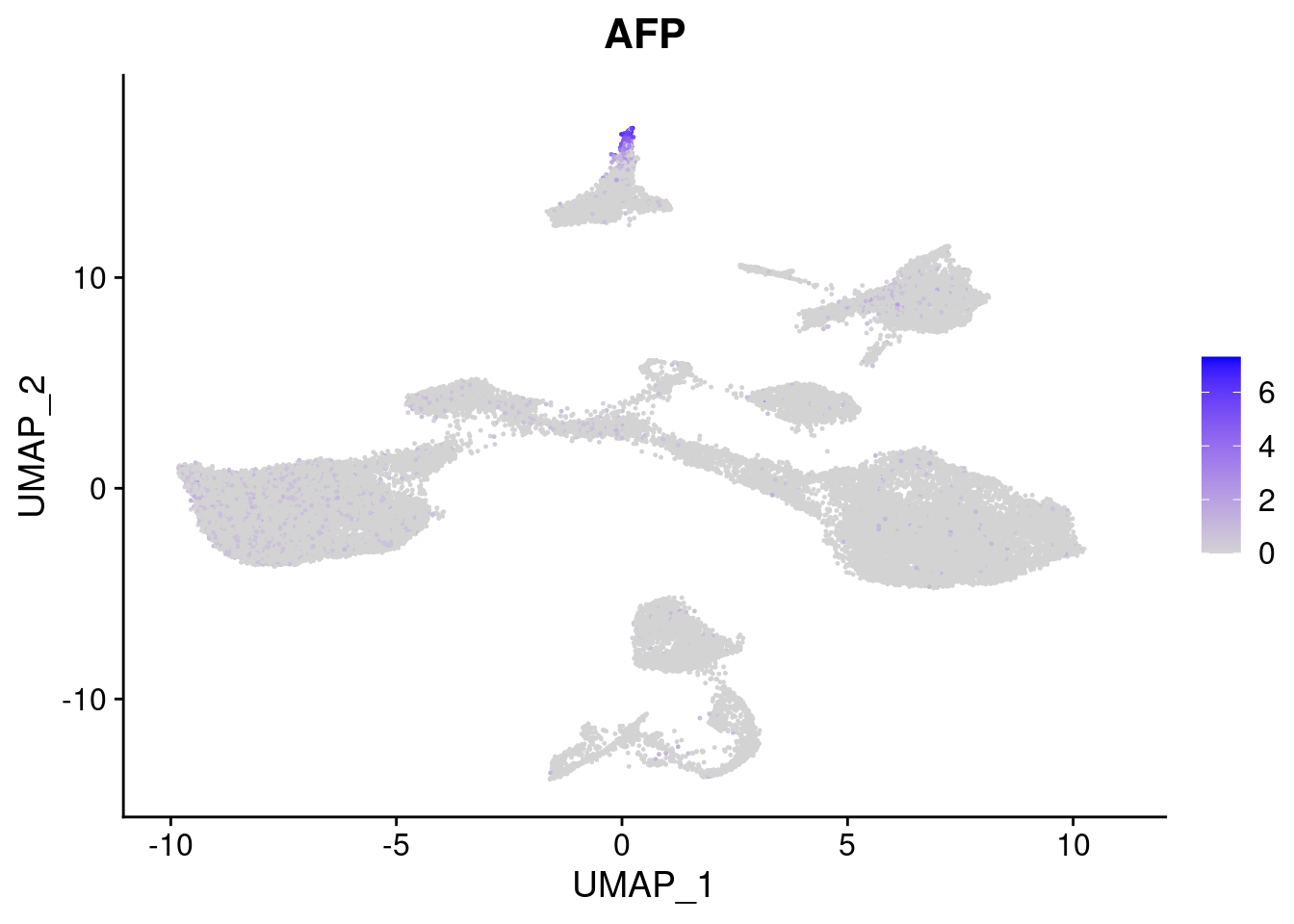
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
[[6]]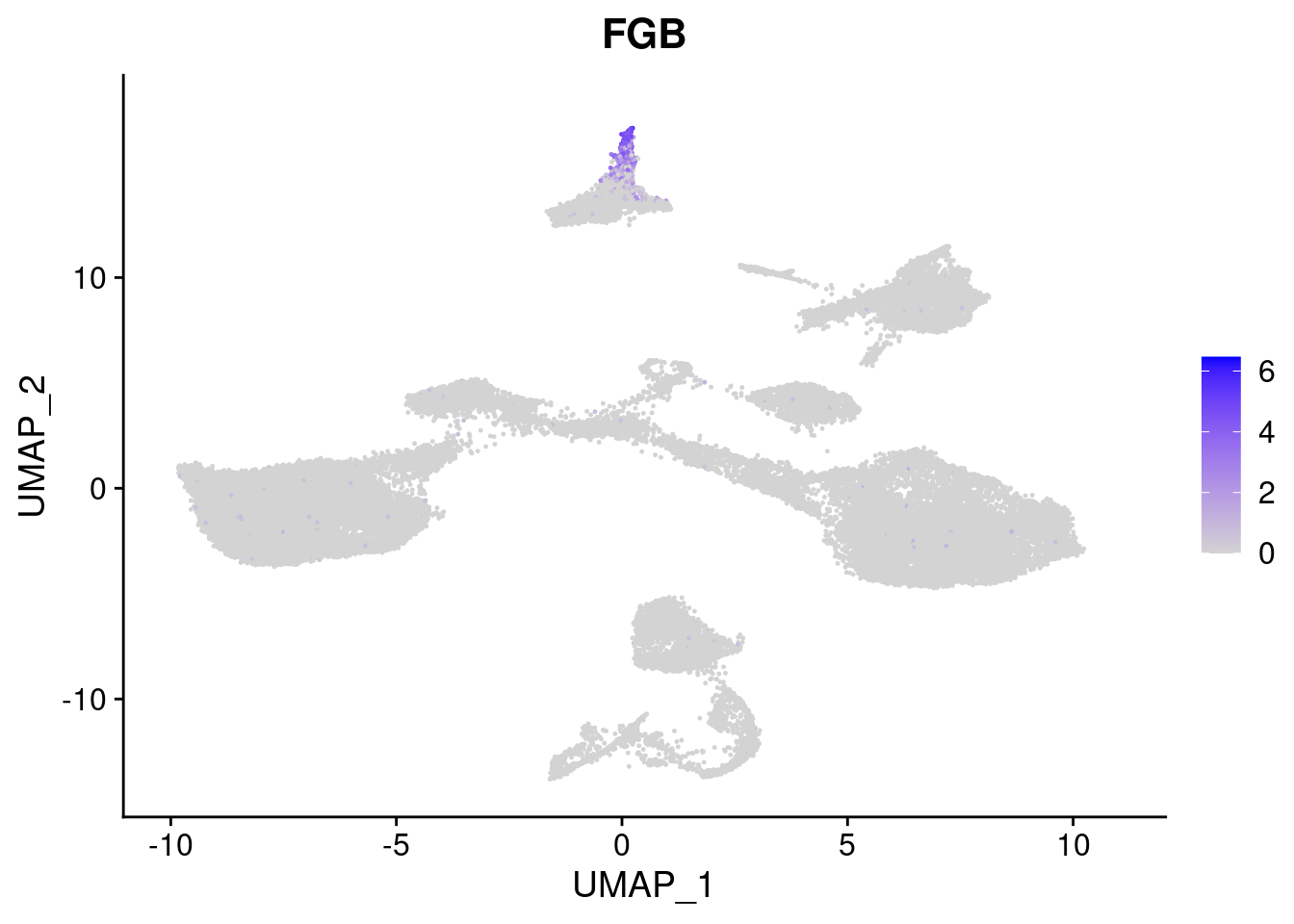
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
#Mesoderm Markers (first 3 early meso markers, 4-6 heart markers, 7-9 endothelial markers (which comes from mesoderm), then some other general muscle markers)
FeaturePlot(merged, features = c("HAND1", "BMP4", "TNNT2","KDR", "GNG11", "ECSCR", "COL3A1", "ACTC1"), pt.size = 0.2, combine=F)[[1]]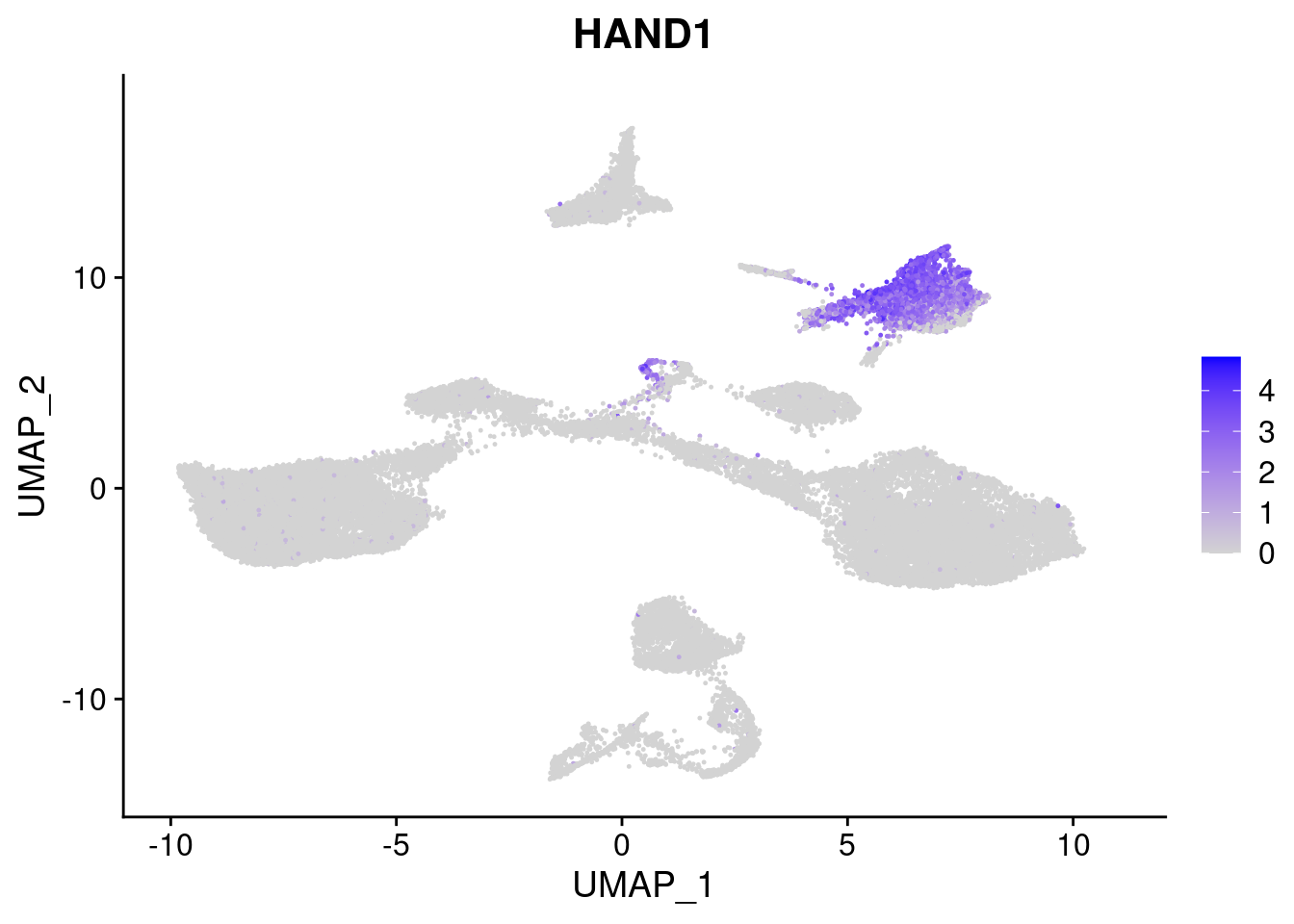
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
[[2]]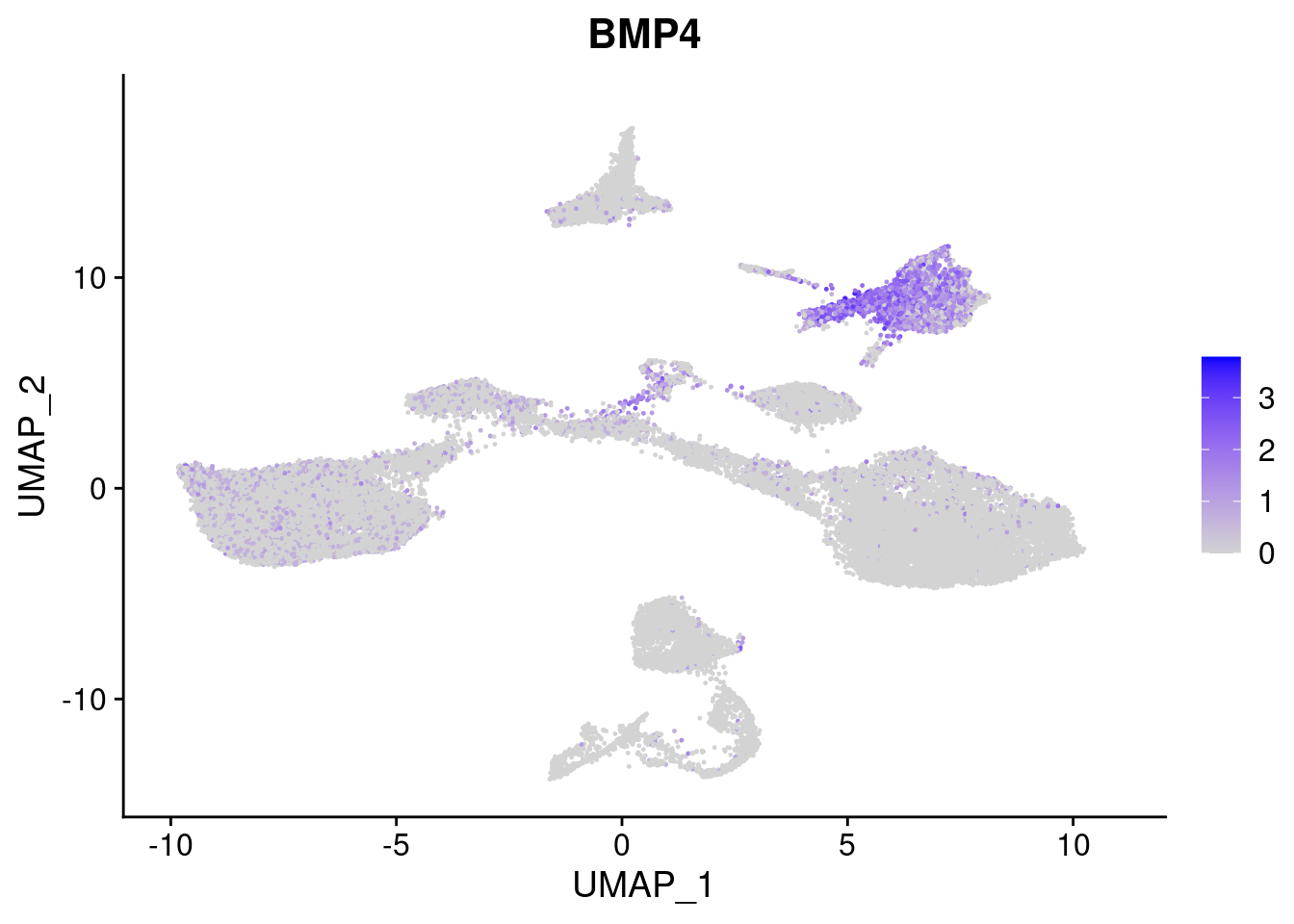
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
[[3]]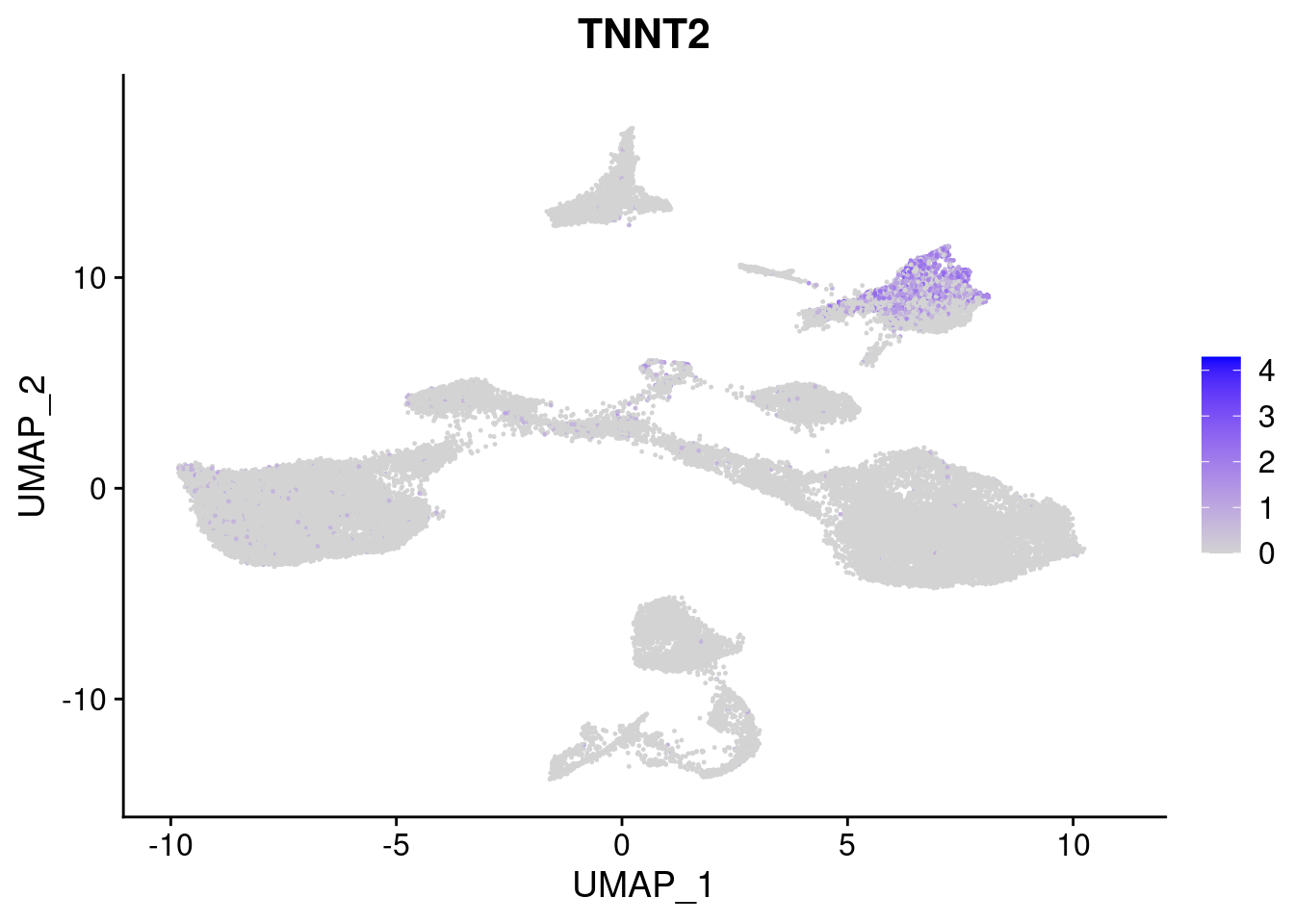
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
[[4]]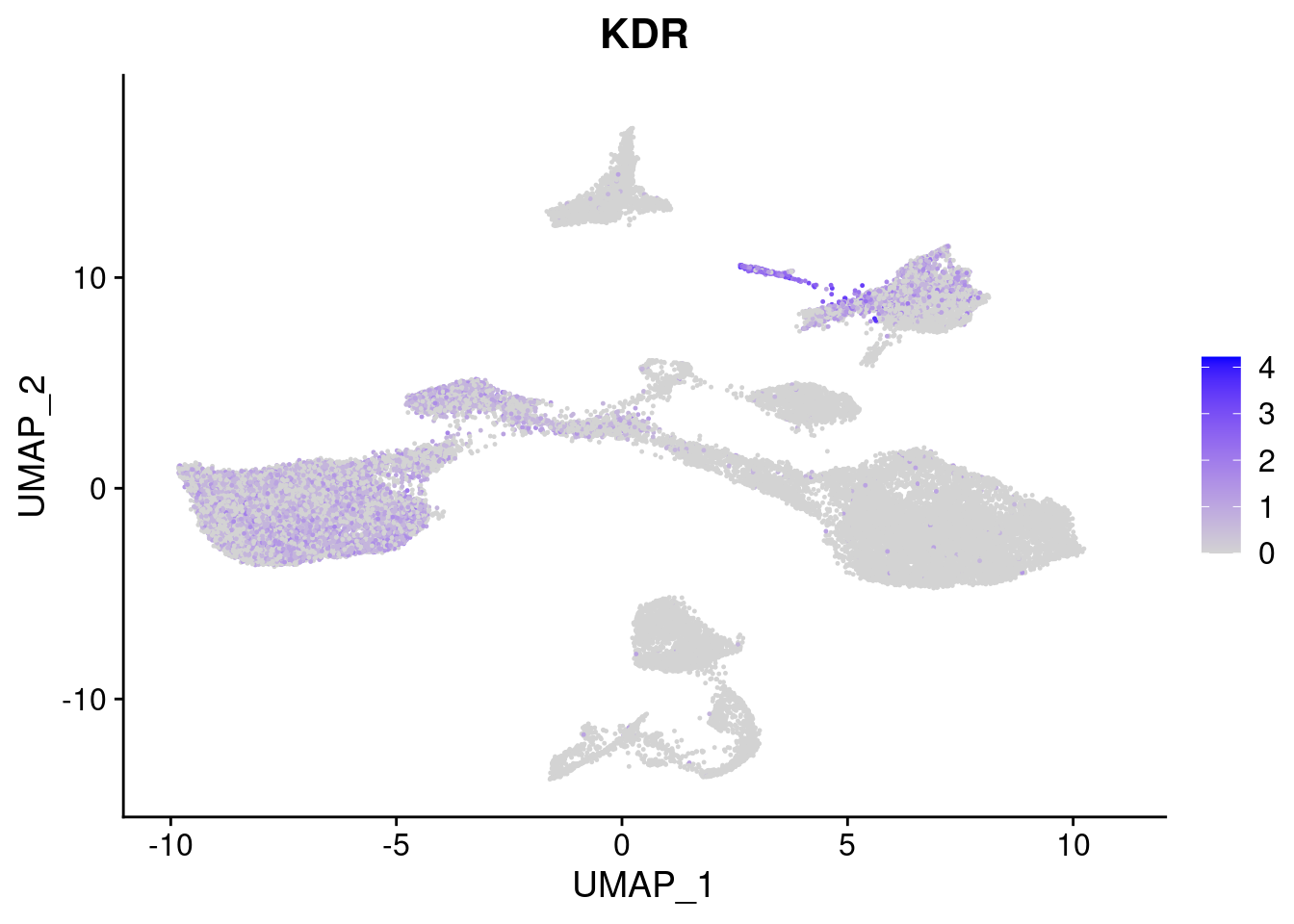
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
[[5]]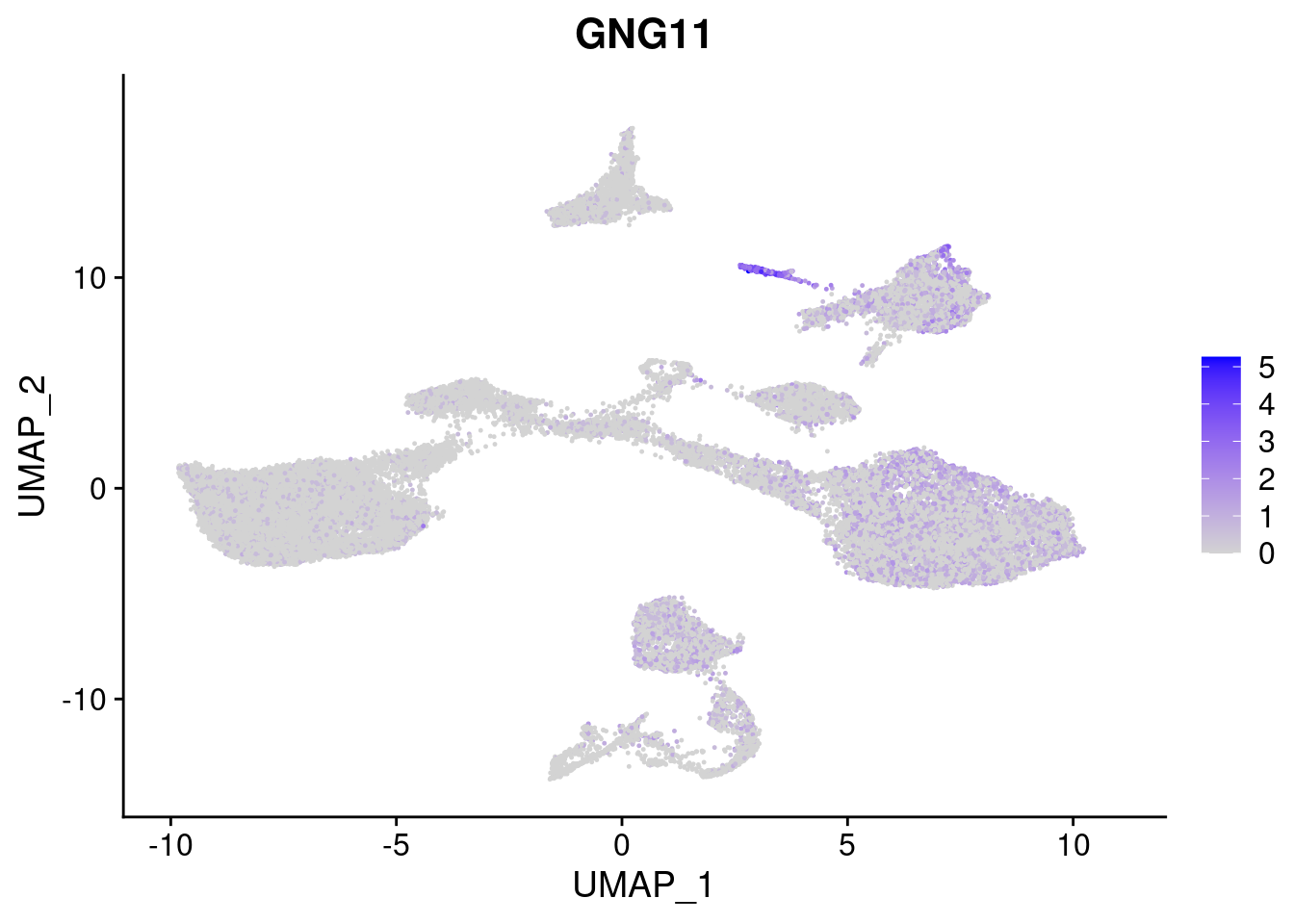
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
[[6]]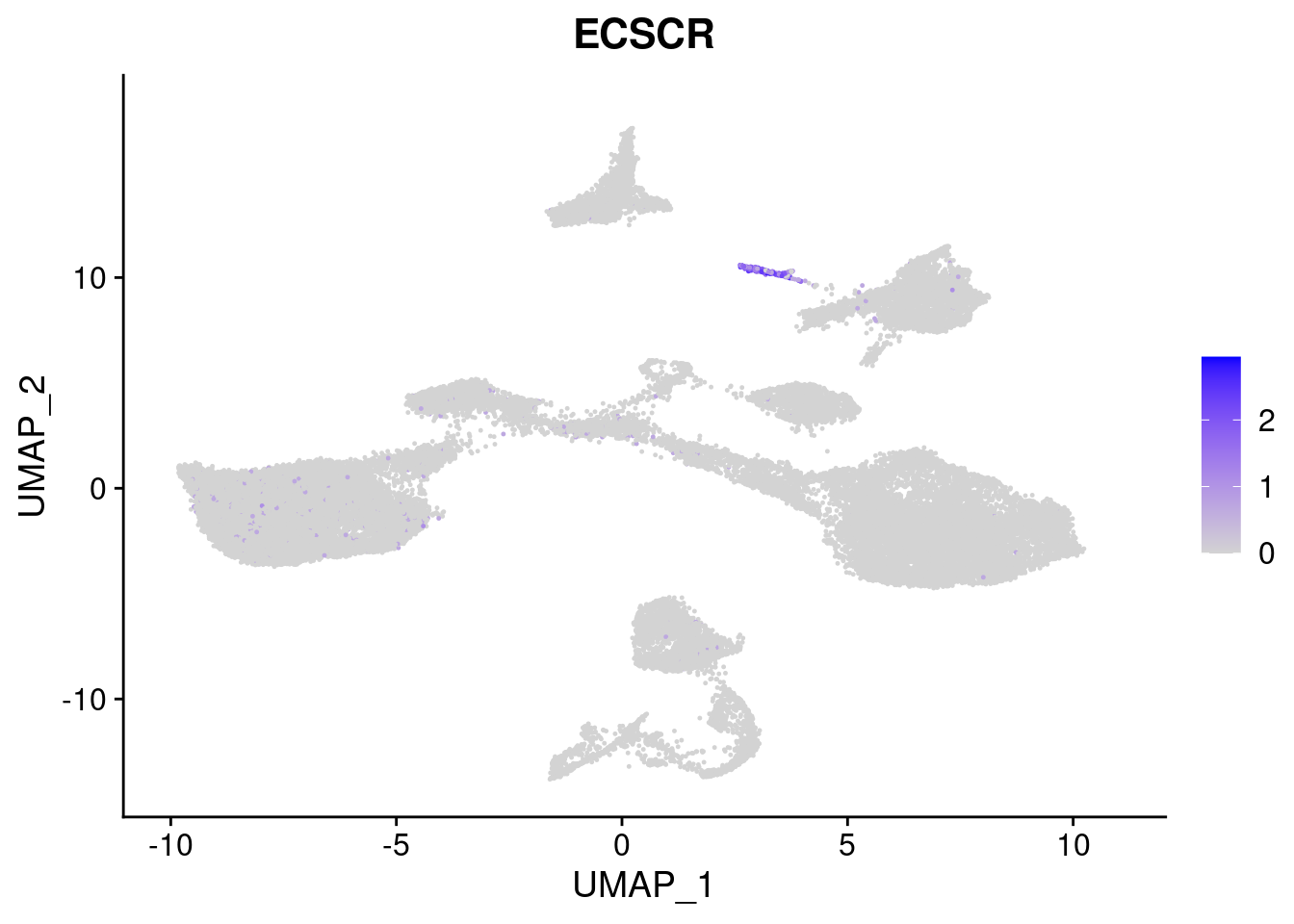
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
[[7]]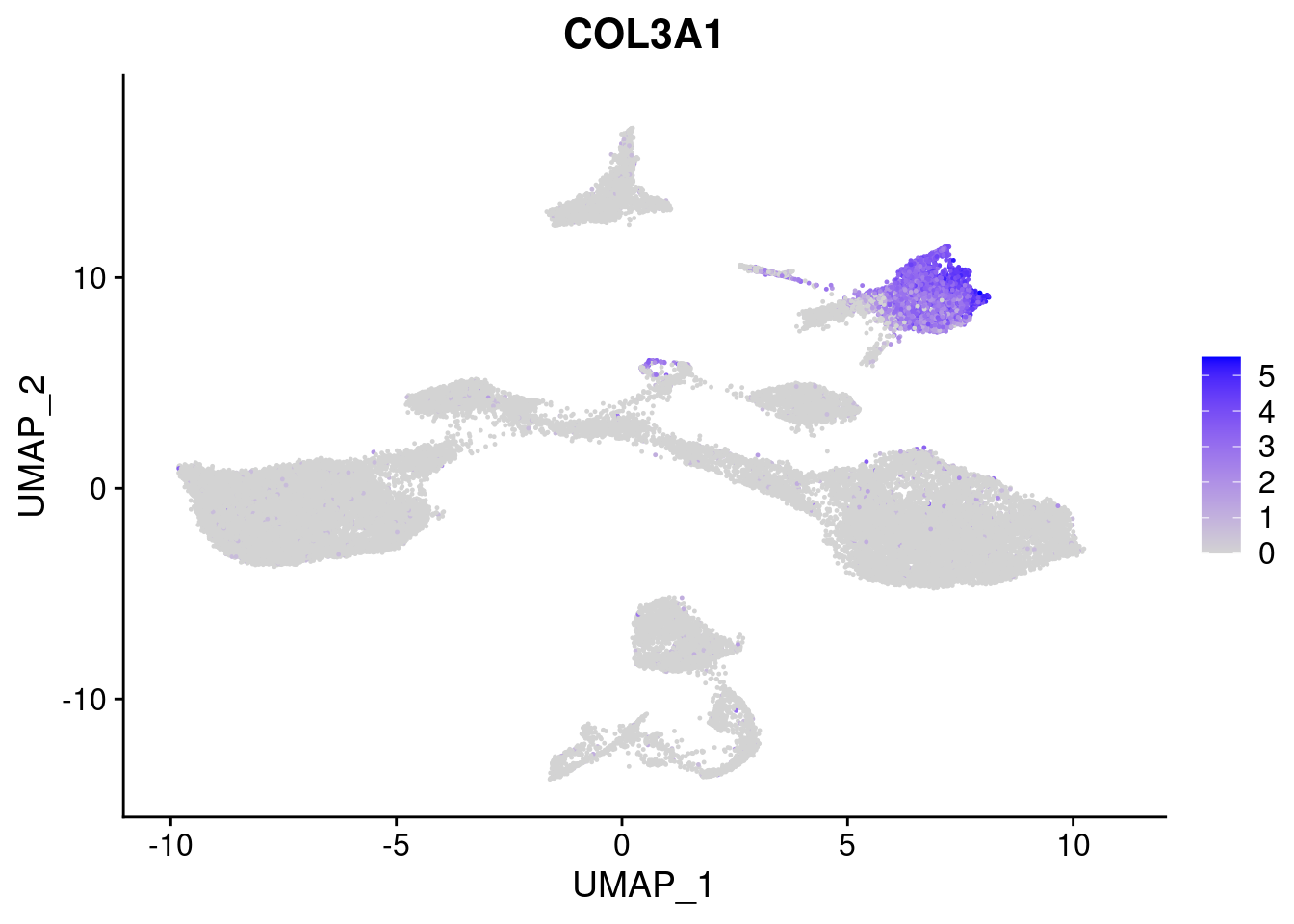
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
[[8]]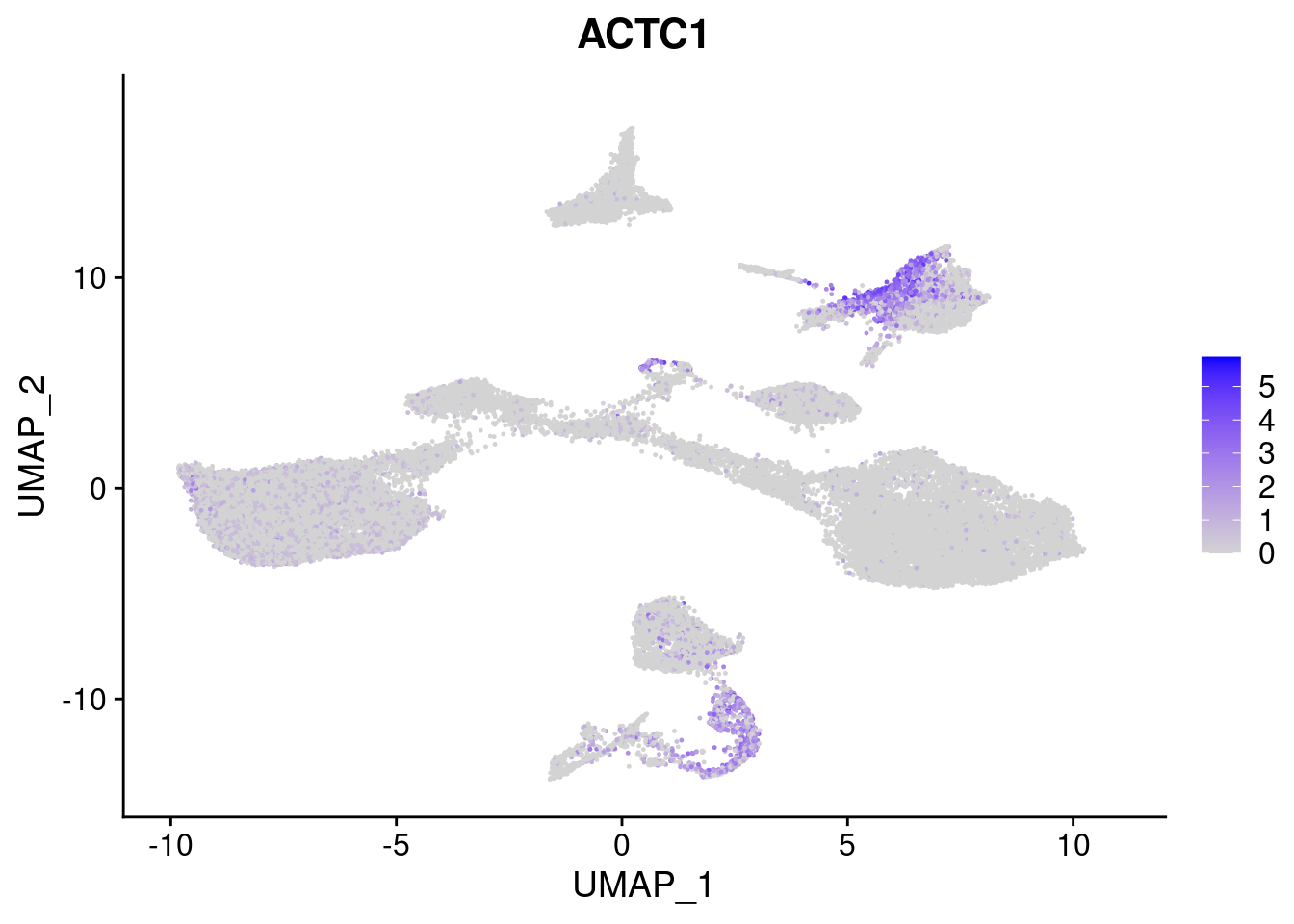
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
#Ectoderm Markers (3-1 early ectoderm markers, 4-6schwann cell (myelinating, non myelinating, or precursor), 7-8 oligodendrocytes, 9-10 radial glia)
FeaturePlot(merged, features = c("PAX6", "GBX2", "NES", "MPZ", "SOX10","GAP43", "OLIG1", "OLIG2", "VIM", "HES5"), pt.size = 0.2, ncol=3, combine=F)[[1]]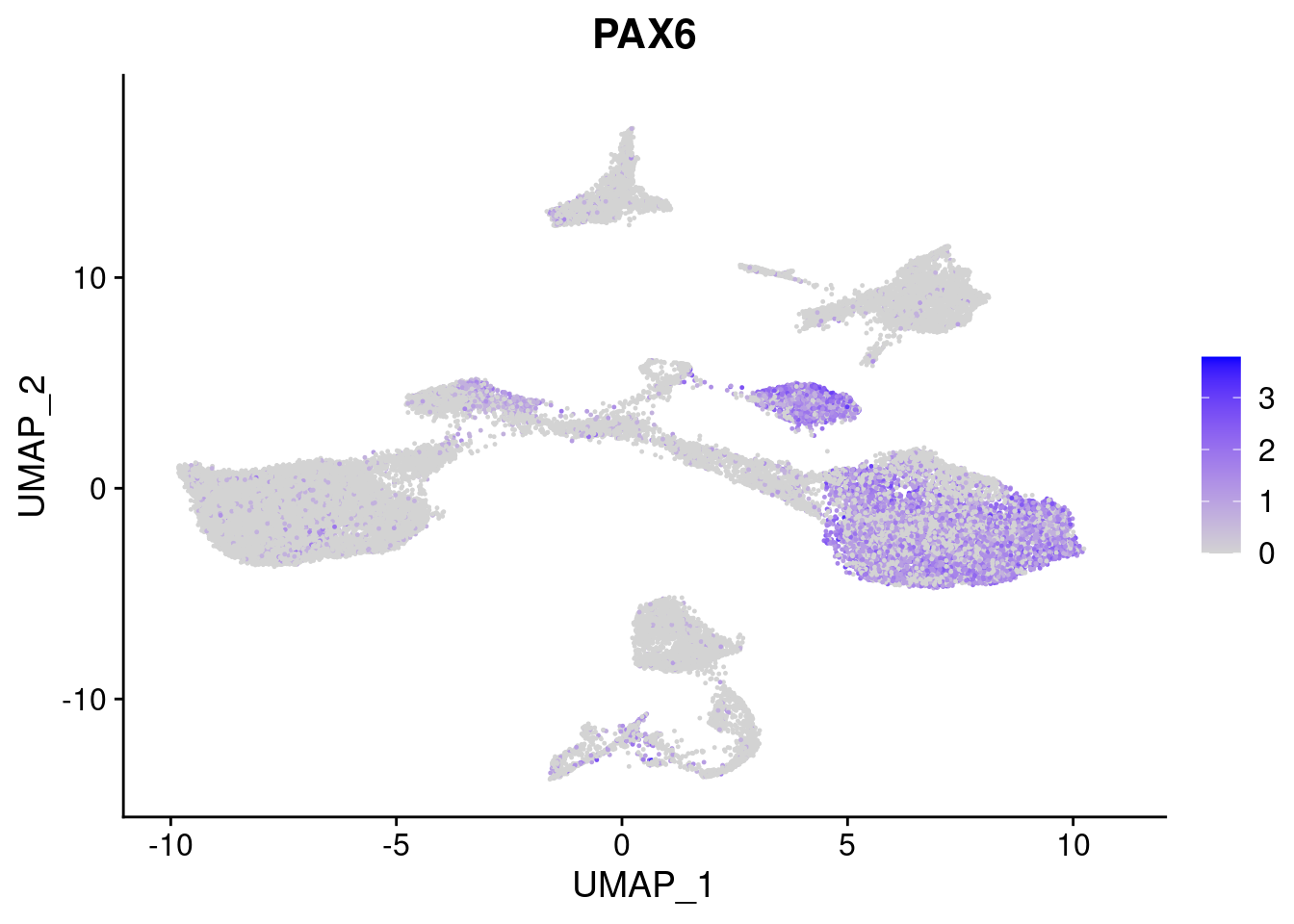
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
[[2]]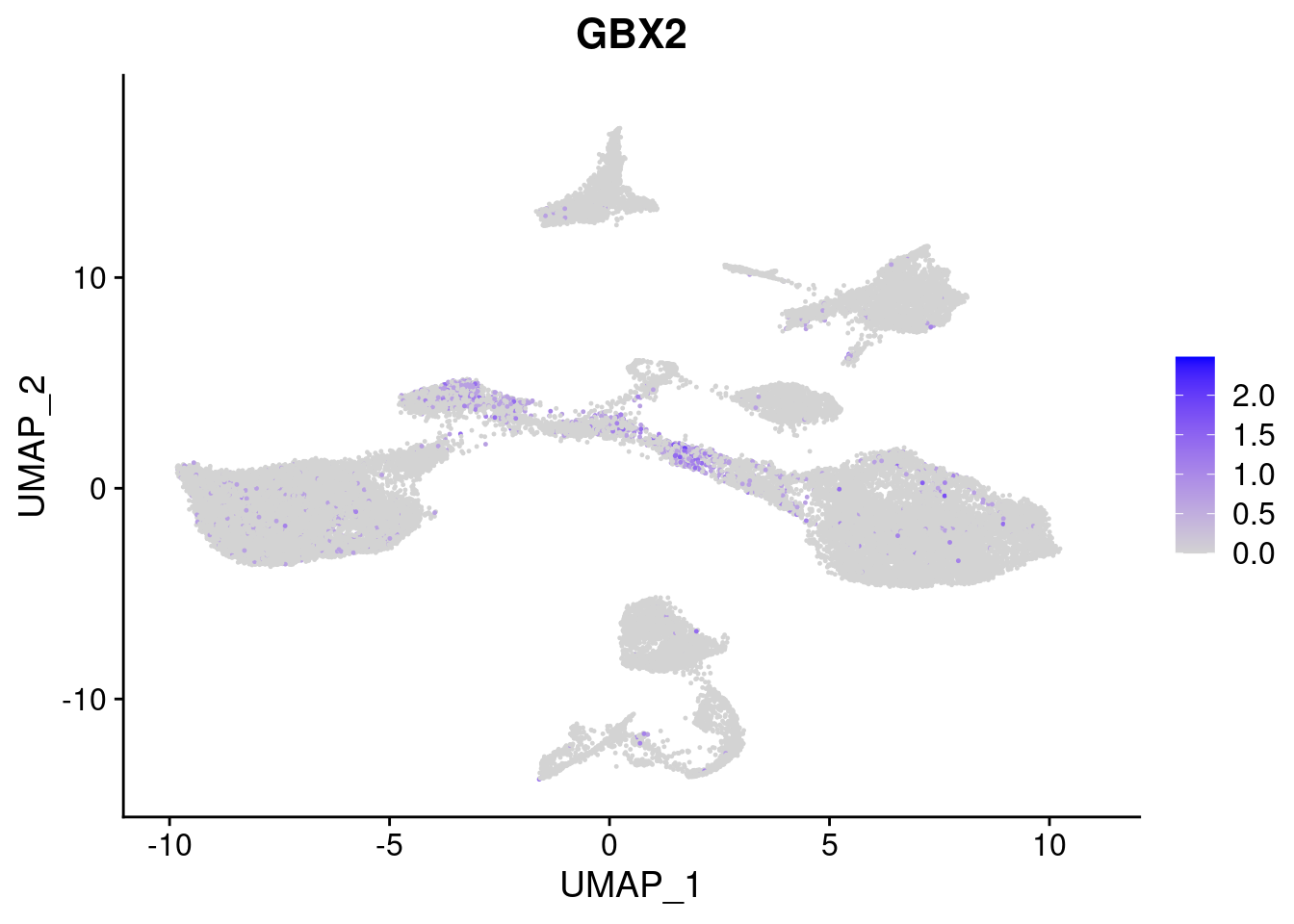
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
[[3]]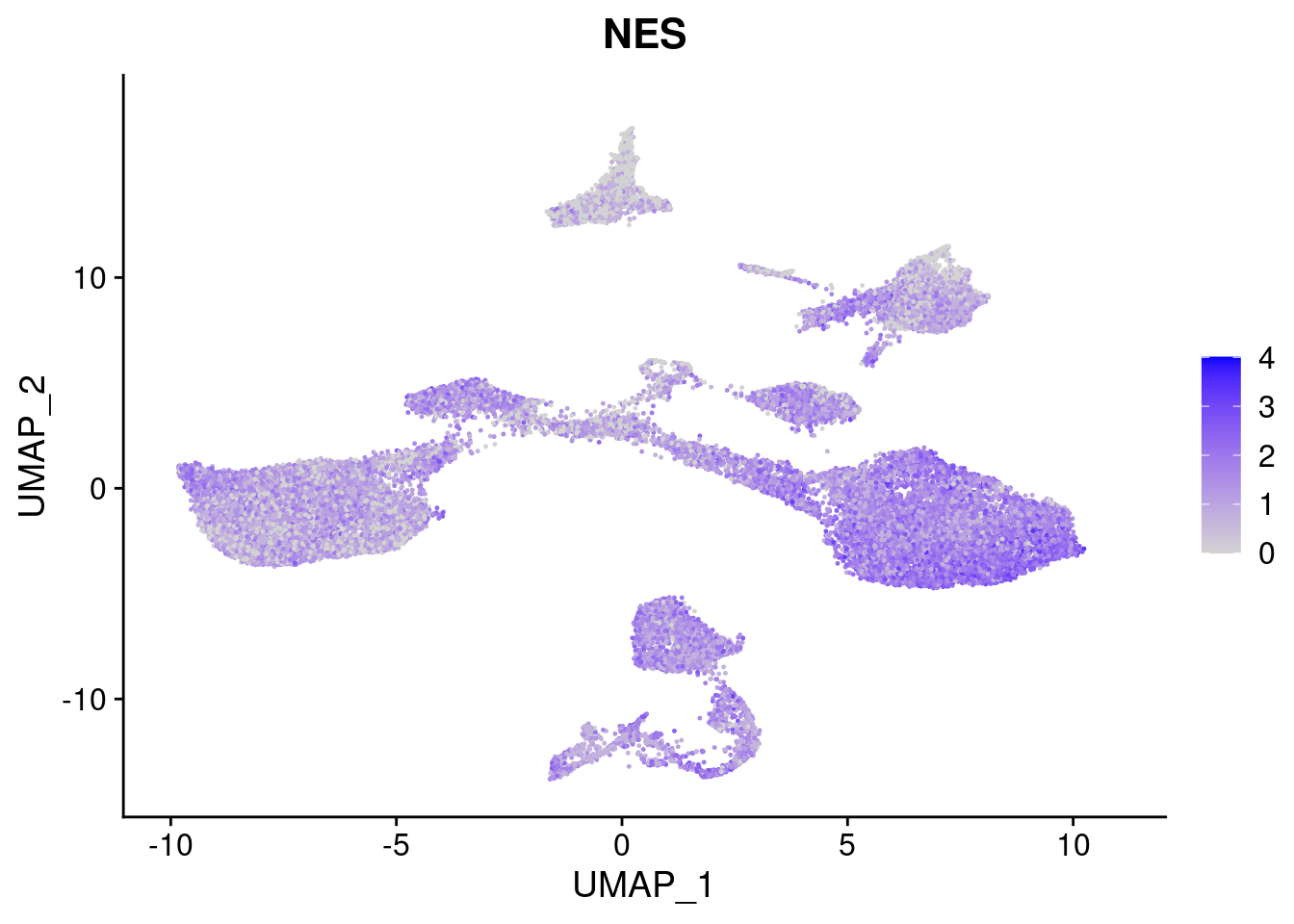
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
[[4]]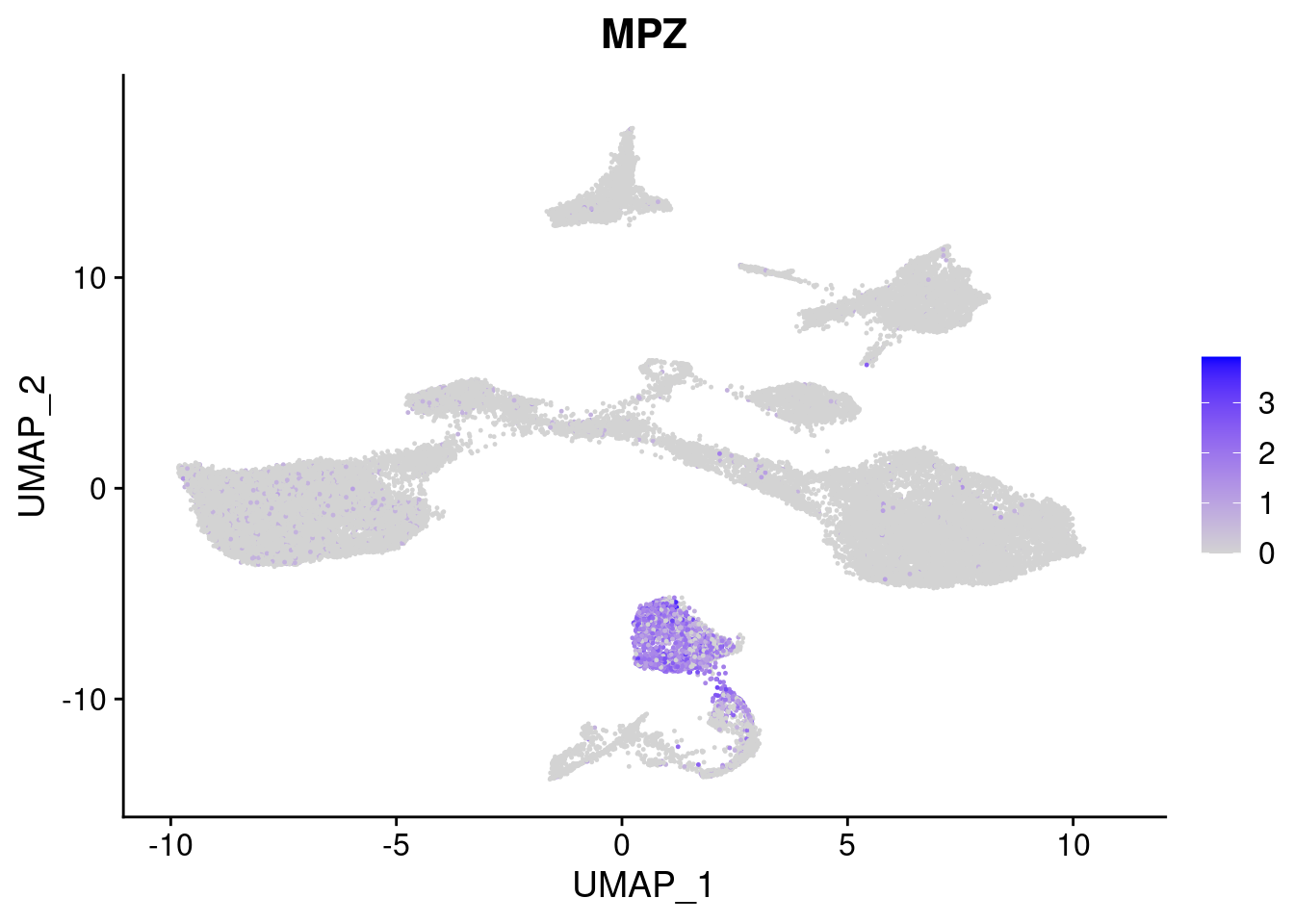
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
[[5]]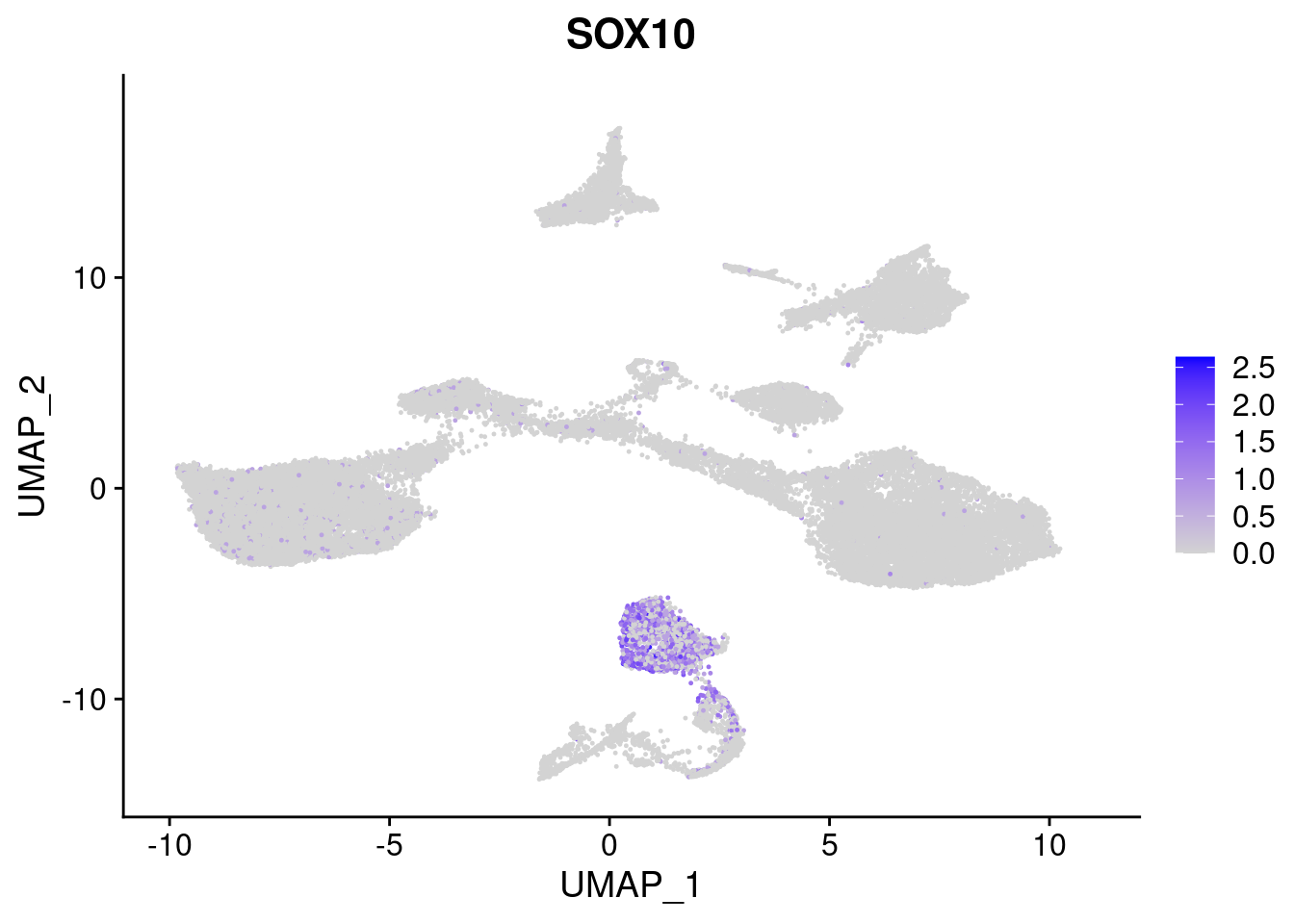
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
[[6]]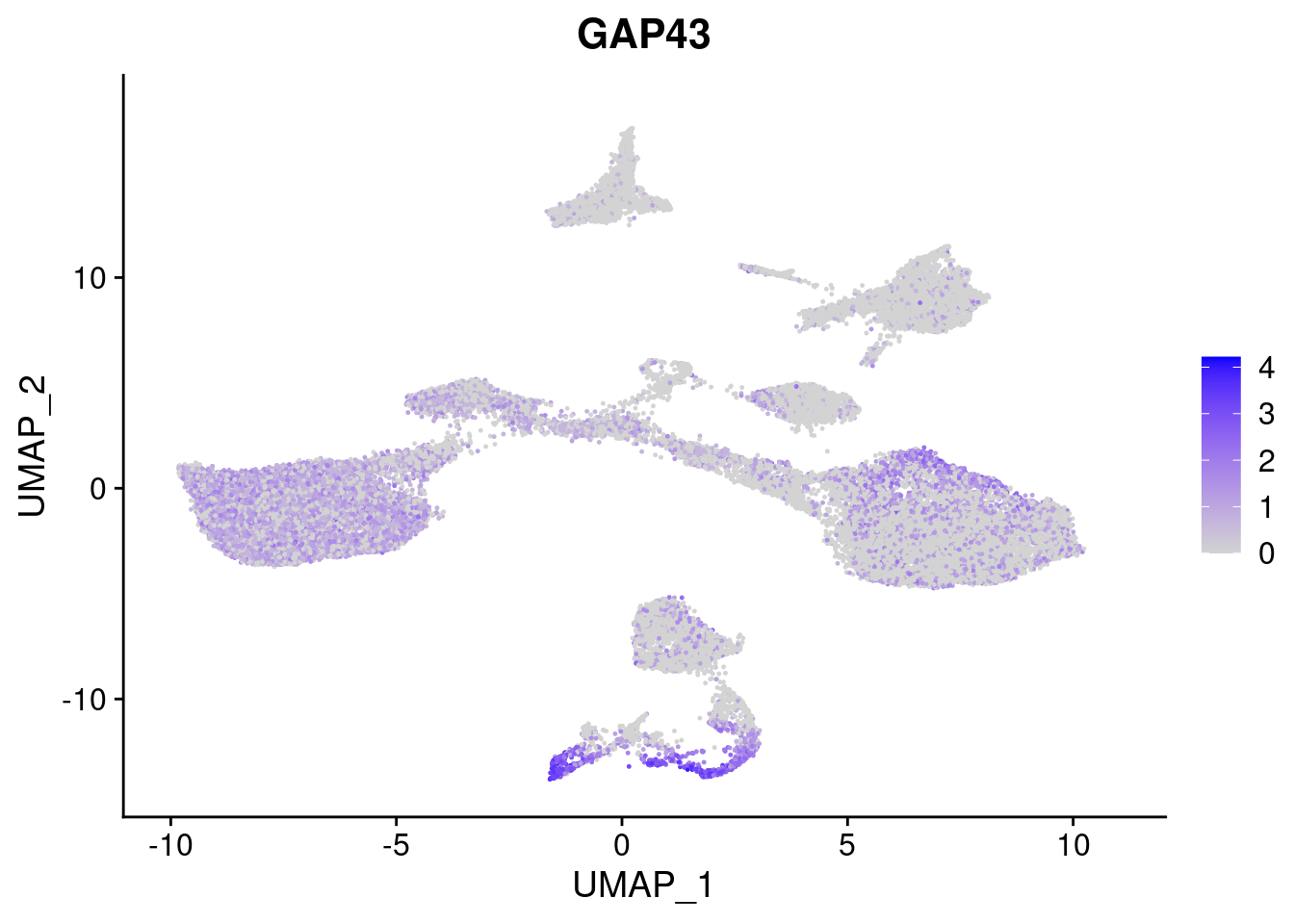
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
[[7]]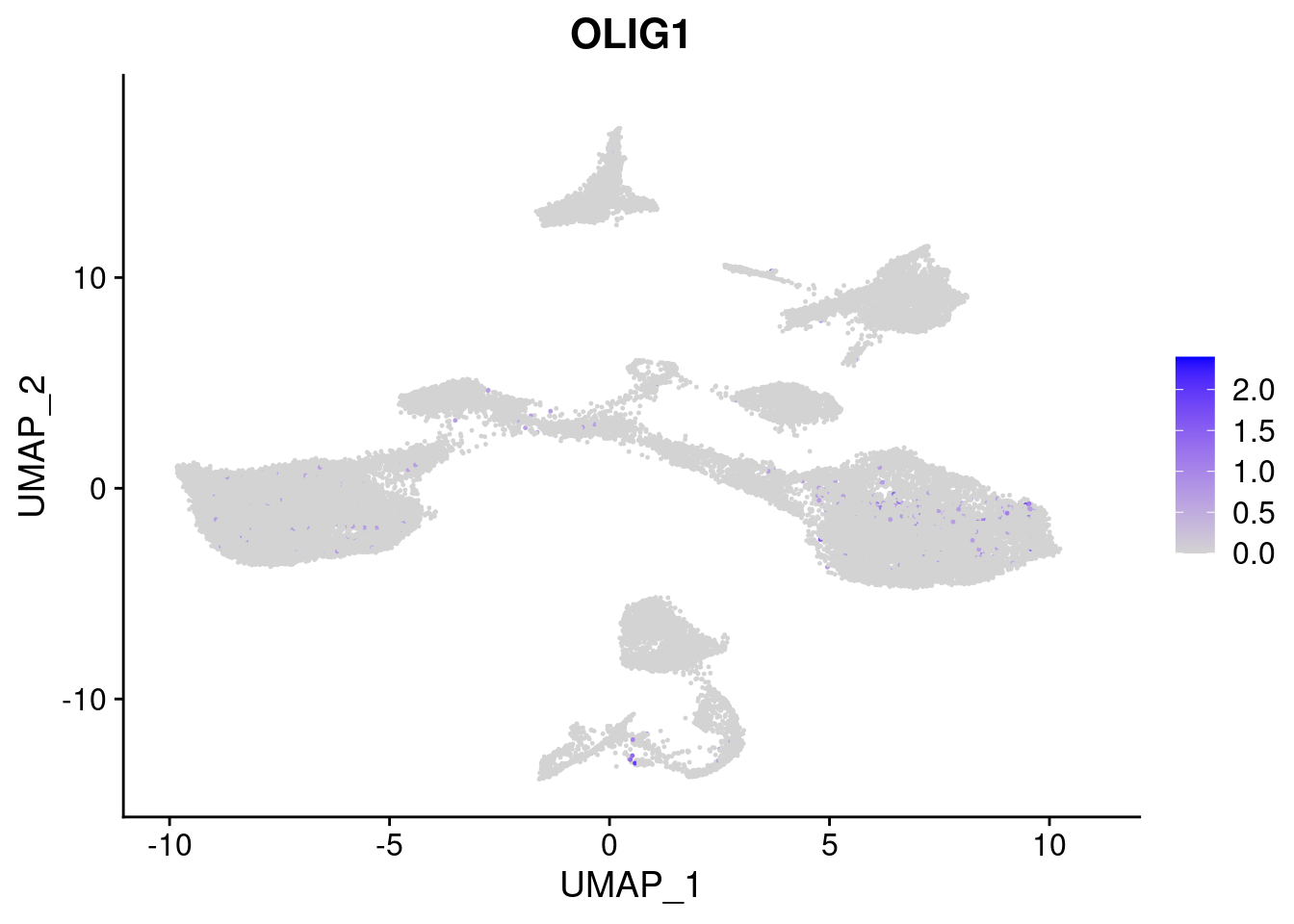
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
[[8]]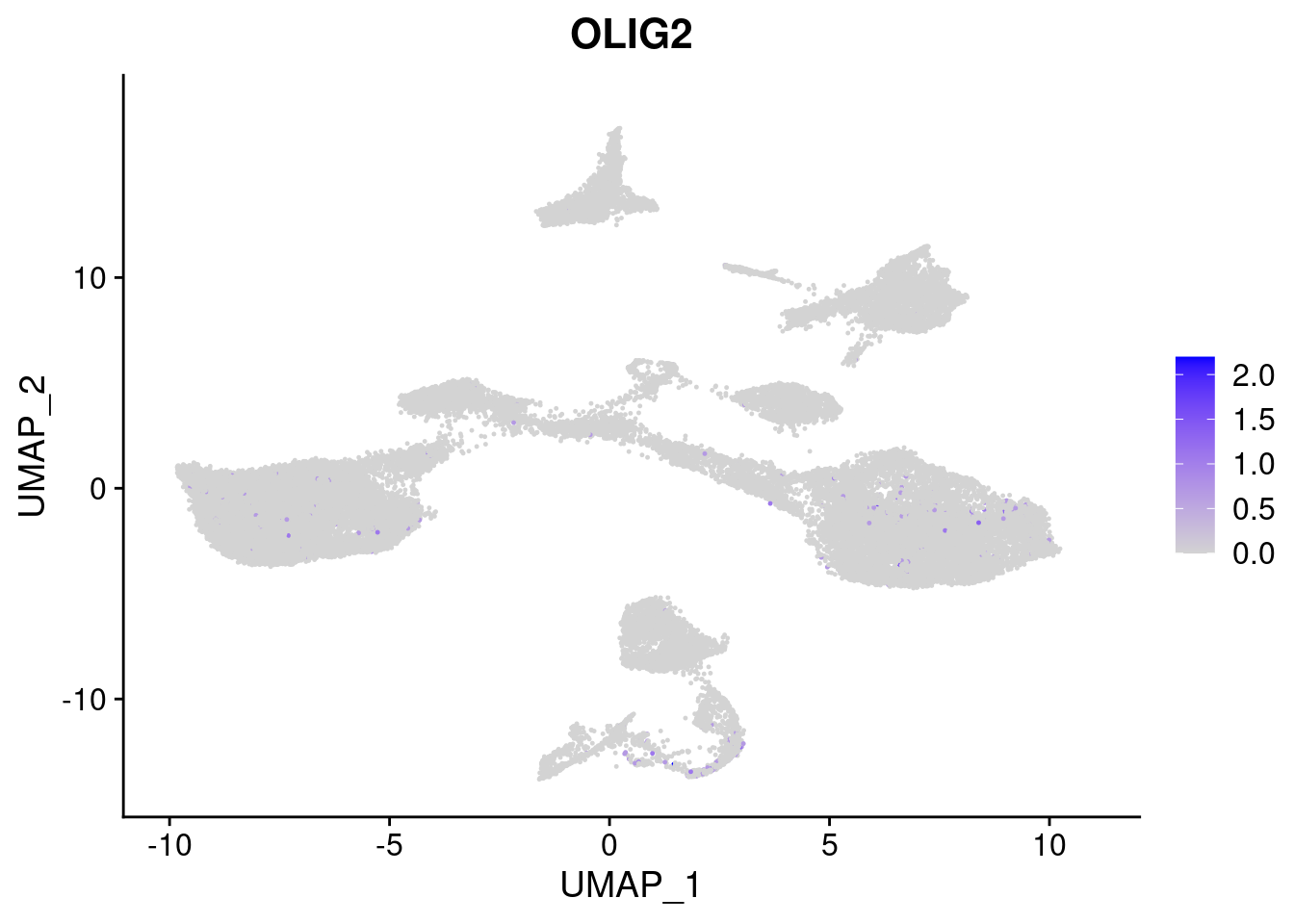
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
[[9]]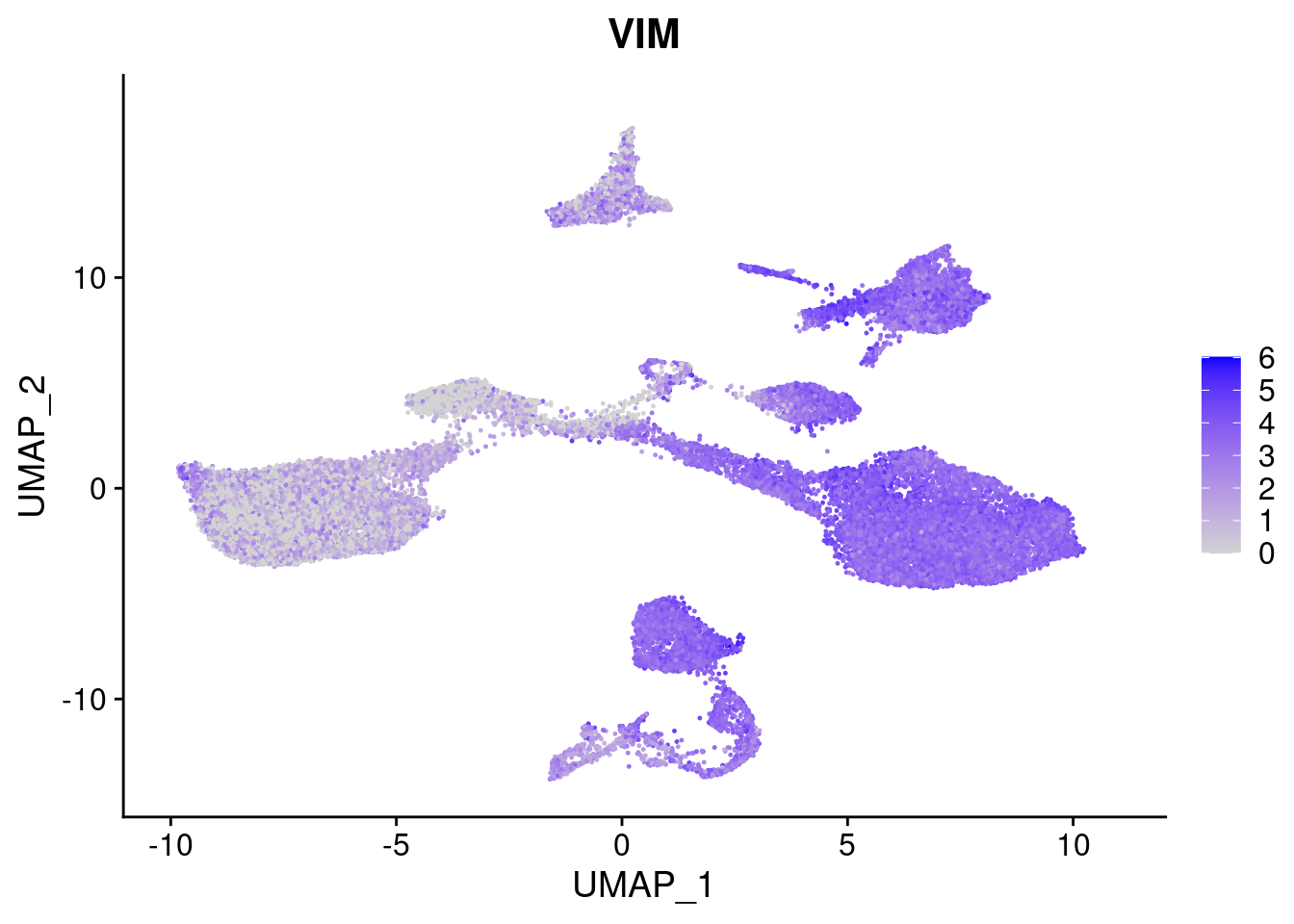
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
[[10]]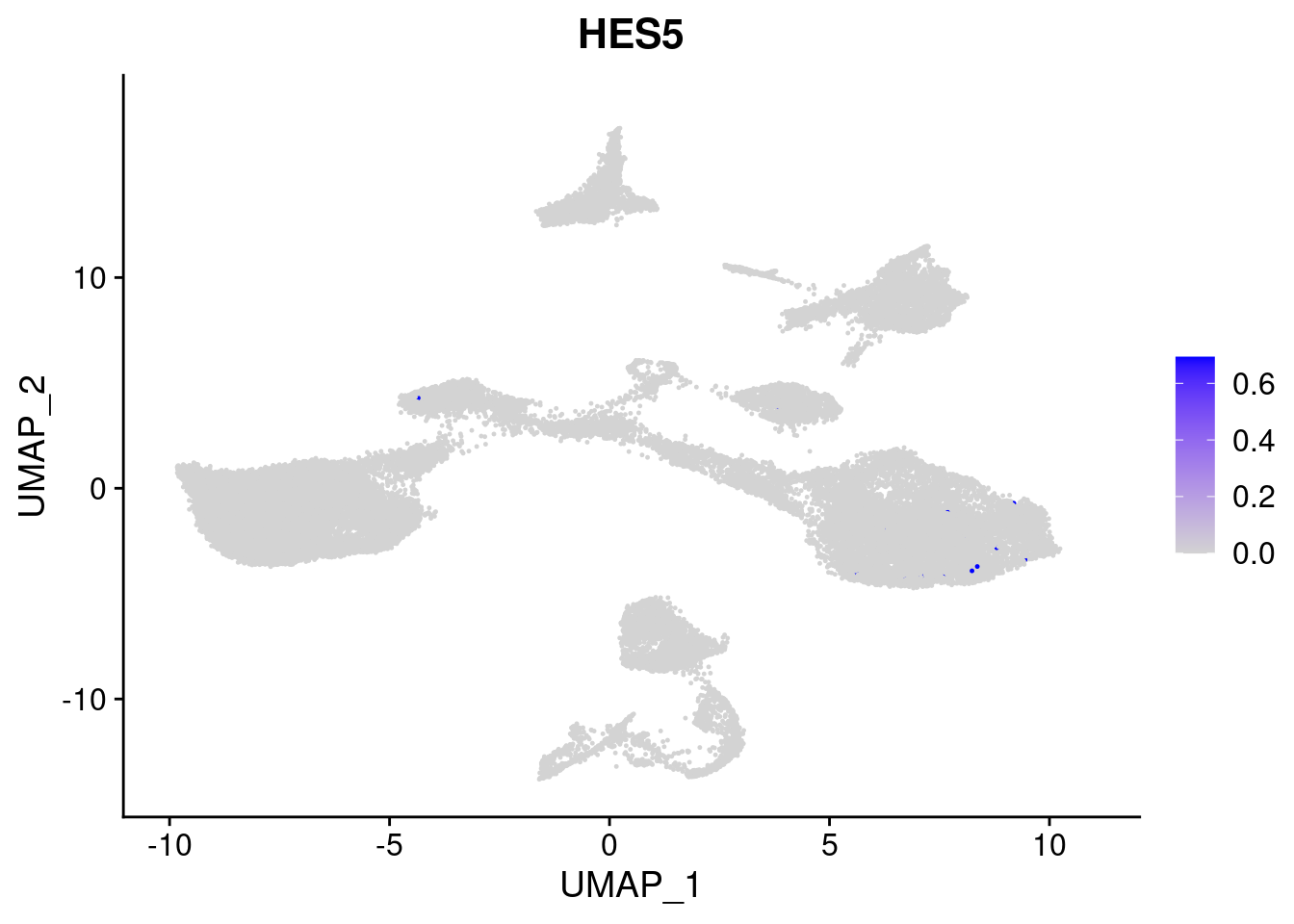
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
#More ectoderm, specifically neurons
#immature neurons: NEUROD1
#Mature Neurons: MAP2, SYP
#dopaminergic: TH, FOXA2,
FeaturePlot(merged, features = c("MAP2", "SYP","NEUROD1", "TH" ), pt.size = 0.2, ncol=3)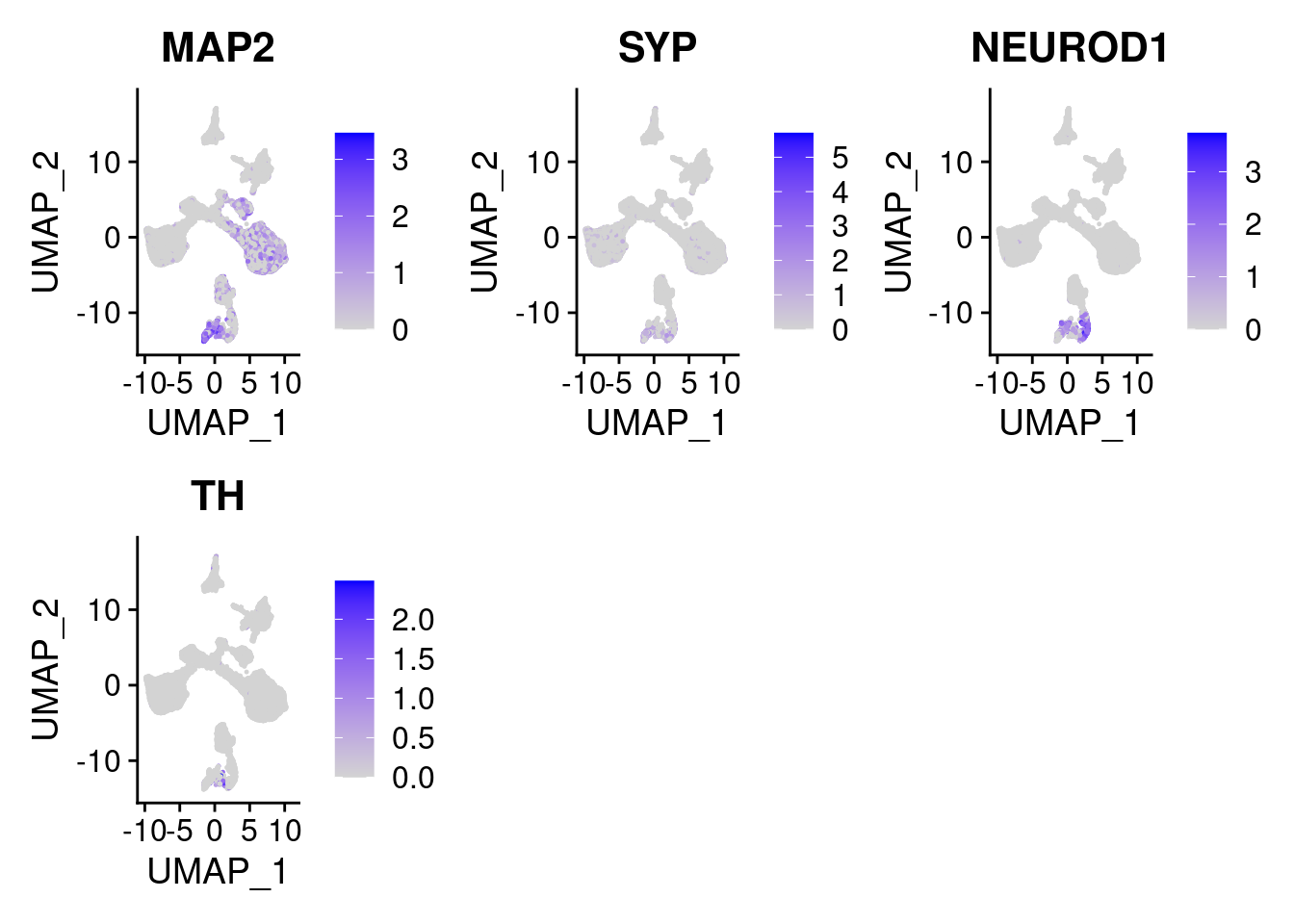
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
Identify cluster markers
#how many cells per cluster?
t1<-table(merged@meta.data$SCT_snn_res.1, merged@meta.data$Batch)
t1
Batch1 Batch2 Batch3
0 2354 1403 1806
1 1858 1022 1482
2 1730 1219 1248
3 1435 503 1131
4 1435 517 652
5 478 885 719
6 827 329 707
7 86 118 1377
8 741 210 509
9 313 227 857
10 246 275 847
11 496 646 224
12 536 404 379
13 130 266 887
14 458 400 270
15 586 185 298
16 665 206 167
17 356 129 550
18 443 189 395
19 281 225 250
20 92 113 369
21 212 185 166
22 510 4 0
23 259 74 109
24 102 60 133
25 100 39 83
26 70 22 104
27 74 10 31#percent of cells in each cluster per batch
t1colsum<- colSums(t1)
percT1<-t1/t1colsum
percT1
Batch1 Batch2 Batch3
0 0.1395128312 0.1422199696 0.1146666667
1 0.1883426254 0.0648888889 0.0878326320
2 0.1098412698 0.0722455995 0.1265078561
3 0.0850471167 0.0509883426 0.0718095238
4 0.1454637608 0.0328253968 0.0386416168
5 0.0303492063 0.0524506608 0.0728839331
6 0.0490132164 0.0333502281 0.0448888889
7 0.0087176888 0.0074920635 0.0816096723
8 0.0470476190 0.0124459195 0.0515965535
9 0.0185503467 0.0230106437 0.0544126984
10 0.0249366447 0.0174603175 0.0501985420
11 0.0314920635 0.0382860191 0.0227065383
12 0.0317667279 0.0409528637 0.0240634921
13 0.0131779017 0.0168888889 0.0525691934
14 0.0290793651 0.0237065134 0.0273694881
15 0.0347300421 0.0187531678 0.0189206349
16 0.0674100355 0.0130793651 0.0098974693
17 0.0226031746 0.0076453506 0.0557526609
18 0.0262549636 0.0191586417 0.0250793651
19 0.0284845413 0.0142857143 0.0148165709
20 0.0058412698 0.0066970900 0.0374049671
21 0.0125644521 0.0187531678 0.0105396825
22 0.0516979219 0.0002539683 0.0000000000
23 0.0164444444 0.0043857050 0.0110491637
24 0.0060451609 0.0060821085 0.0084444444
25 0.0101368474 0.0024761905 0.0049191015
26 0.0044444444 0.0013038582 0.0105423213
27 0.0043857050 0.0010136847 0.0019682540heatmap(t(percT1))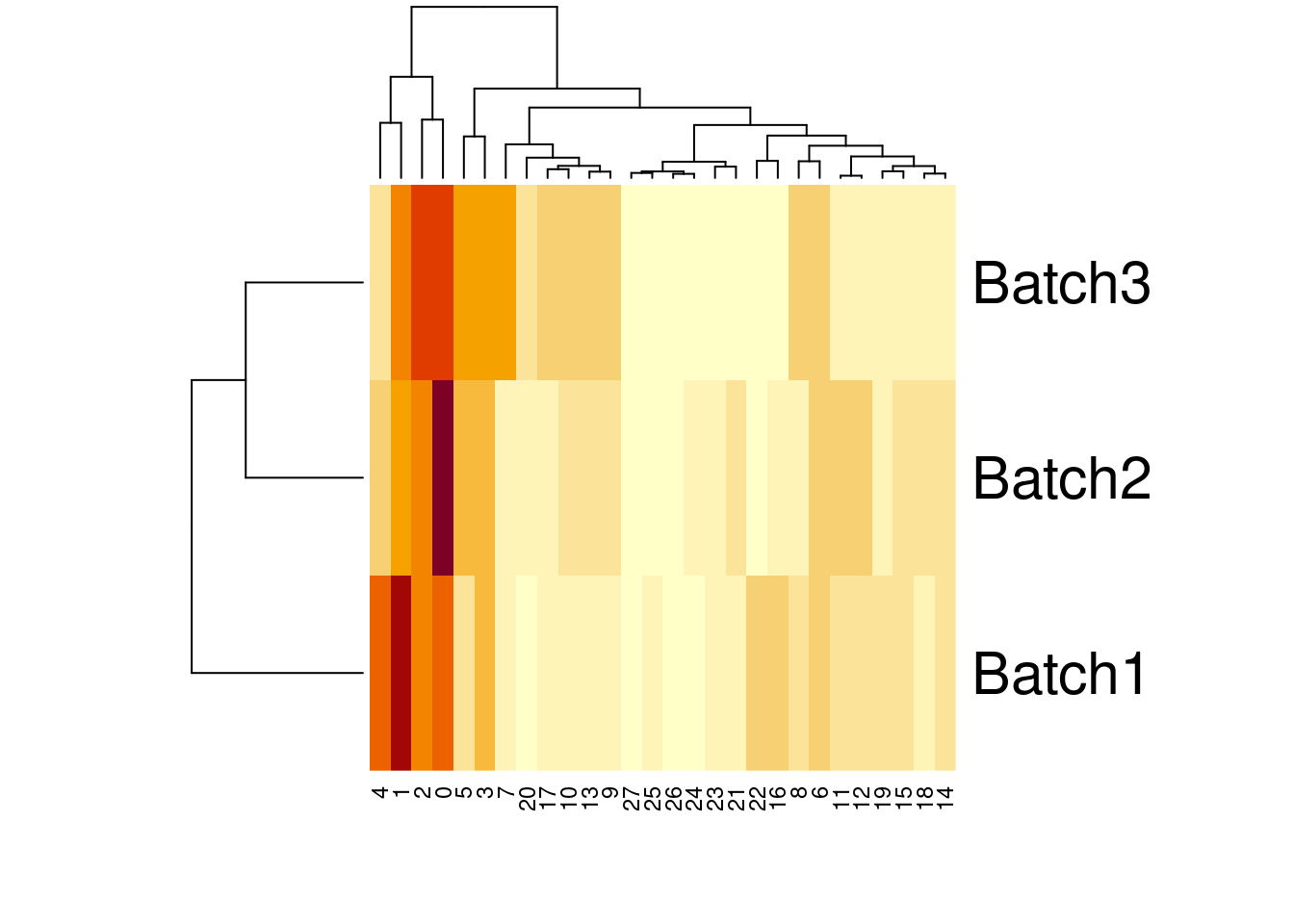
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
#how many cells per cluster from each individual?
t2<-table(merged@meta.data$SCT_snn_res.1, merged@meta.data$individual)
t2
SNG-NA18511 SNG-NA18858 SNG-NA19160
0 4 5556 3
1 4 4358 0
2 4118 29 50
3 946 18 2105
4 438 112 2054
5 12 2061 9
6 21 1839 3
7 1543 12 26
8 135 2 1323
9 740 4 653
10 549 2 817
11 138 1208 20
12 920 21 378
13 154 4 1125
14 82 1004 42
15 76 2 991
16 107 17 914
17 57 5 973
18 138 0 889
19 653 8 95
20 138 0 436
21 408 8 147
22 331 10 173
23 87 169 186
24 89 2 204
25 7 213 2
26 39 6 151
27 15 0 100t2colsums<-colSums(t2)
percT2<- t2/t2colsums
percT2
SNG-NA18511 SNG-NA18858 SNG-NA19160
0 0.0003347560 0.3332933413 0.0002163098
1 0.0002399520 0.3142259716 0.0000000000
2 0.2969211911 0.0024269813 0.0029994001
3 0.0791698050 0.0010797840 0.1517773452
4 0.0262747451 0.0080755642 0.1718972299
5 0.0008652390 0.1724830530 0.0005398920
6 0.0017574692 0.1103179364 0.0002163098
7 0.0925614877 0.0008652390 0.0021759143
8 0.0097339390 0.0001673780 0.0793641272
9 0.0619298686 0.0002399520 0.0470834235
10 0.0329334133 0.0001442065 0.0683739225
11 0.0099502488 0.1010963261 0.0011997600
12 0.0769938907 0.0012597481 0.0272550292
13 0.0092381524 0.0002884130 0.0941501381
14 0.0059124667 0.0840237677 0.0025194961
15 0.0063603649 0.0001199760 0.0714543226
16 0.0064187163 0.0012257553 0.0764917566
17 0.0041098854 0.0004184451 0.0583683263
18 0.0115490836 0.0000000000 0.0640997909
19 0.0391721656 0.0005768260 0.0079504561
20 0.0099502488 0.0000000000 0.0261547690
21 0.0341451167 0.0004799040 0.0105991780
22 0.0198560288 0.0007210325 0.0144781990
23 0.0062729829 0.0141434430 0.0111577684
24 0.0074483220 0.0001199760 0.0147090634
25 0.0004199160 0.0153579926 0.0001673780
26 0.0028120268 0.0005021341 0.0090581884
27 0.0012553352 0.0000000000 0.0072103252heatmap(t(percT2))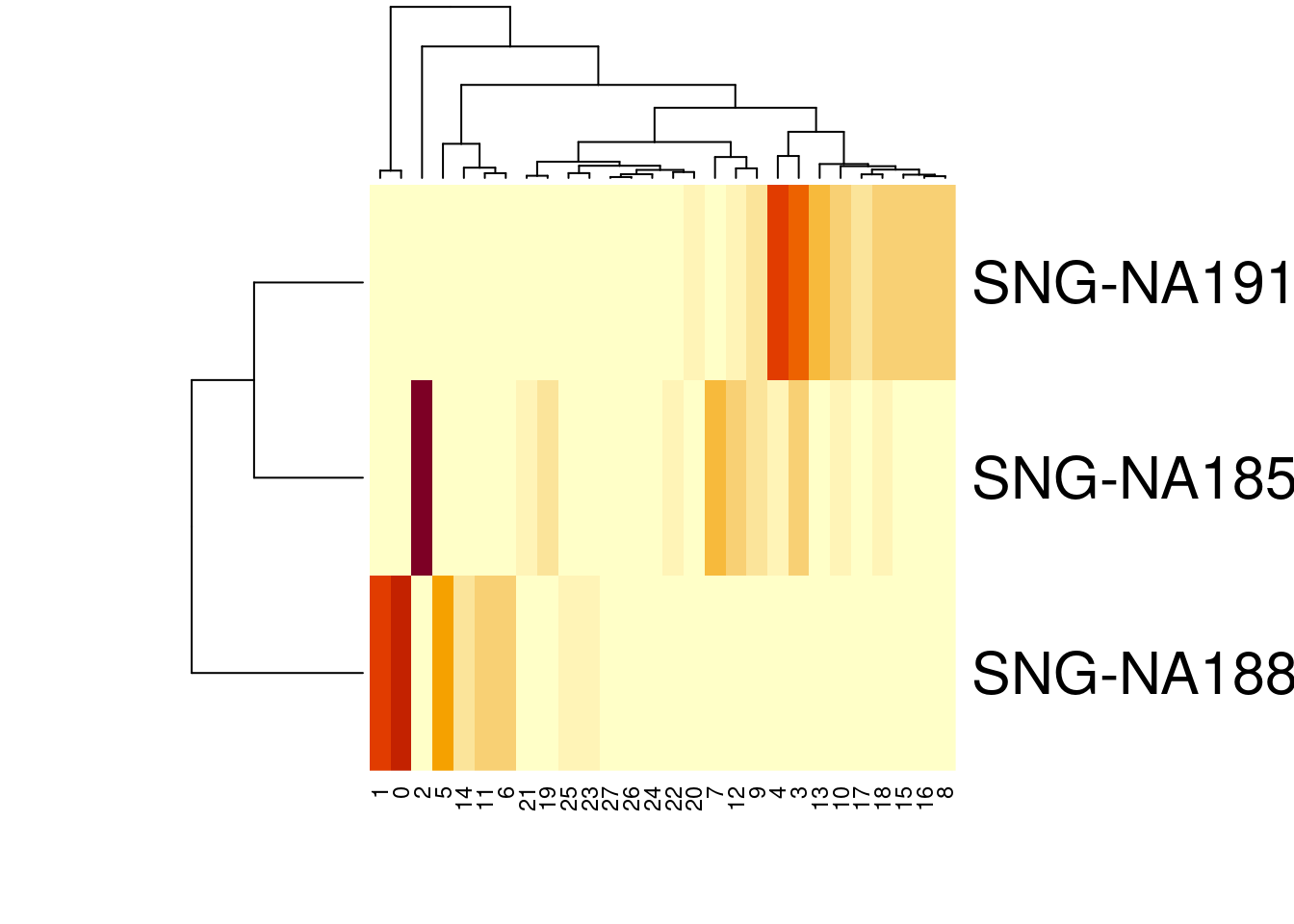
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
cormat<-round(cor(percT2),2)
library(reshape2)
melted_cormat<-melt(cormat)
ugly<-ggplot(data= melted_cormat, aes(x=Var1, y=Var2, fill=value)) +
geom_tile() +
ggtitle("Pairwise Pearson Correlation of the percent of cells from \neach cell line assigned to each Seurat Cluster")
get_lower_tri<- function(cormat){
cormat[upper.tri(cormat)]<-NA
return(cormat)
}
lower_tri<- get_lower_tri(cormat)
melted_tri<- melt(lower_tri)
pretty<-ggplot(data= melted_tri, aes(x=Var1, y=Var2, fill=value)) +
geom_tile(color="white") +
scale_fill_gradient2(low="blue", high="red", mid="white", midpoint = 0, limit= c(-1,1), space= "Lab", name="Pearson\nCorrelation") +
theme_minimal() +
ggtitle("Pairwise Pearson Correlation of the percent of cells from \neach cell line assigned to each Seurat Cluster")
pretty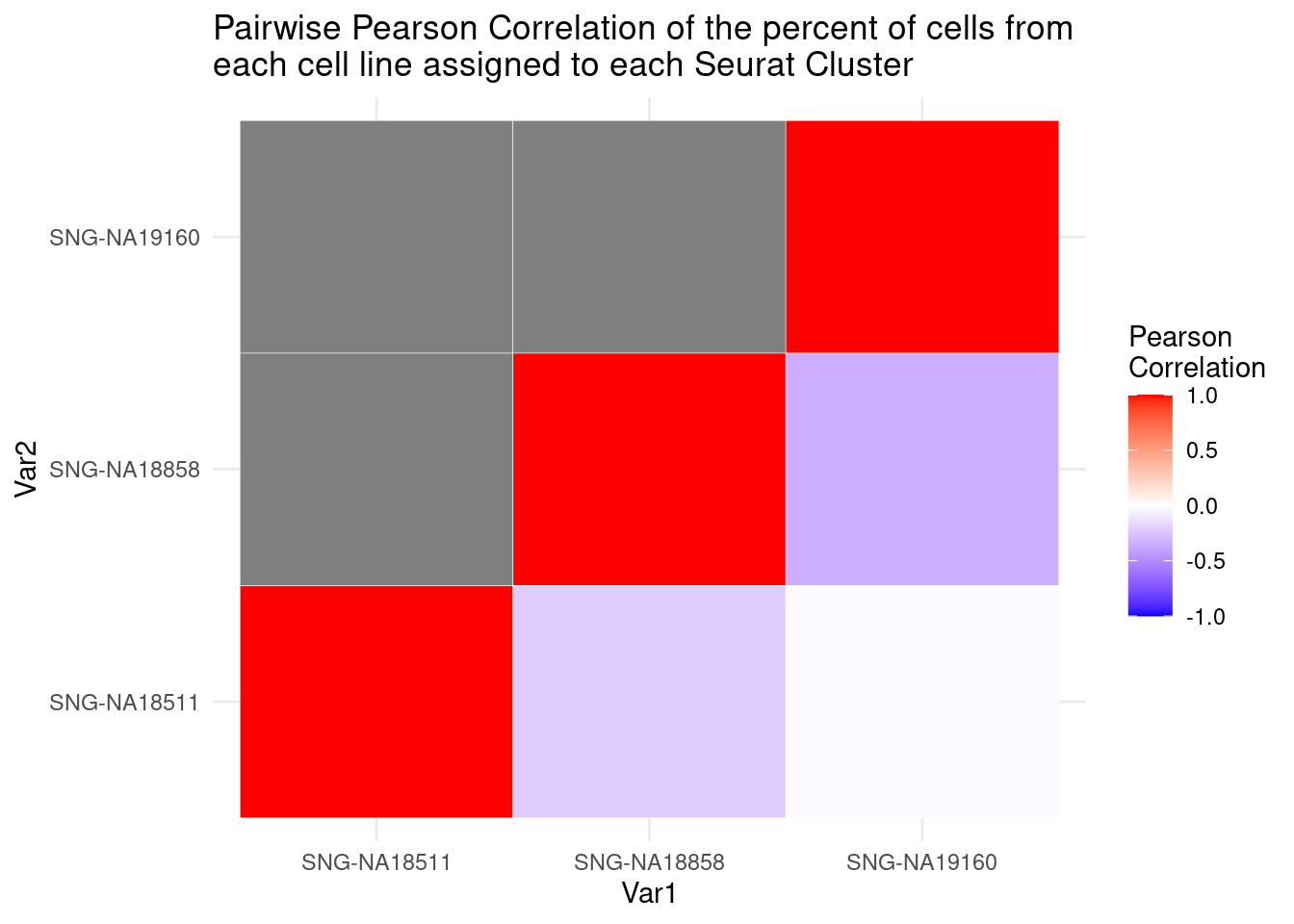
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
#exploring similarity in the number of cells per individual between batches
merged.Batch1<- (subset(merged, Batch == "Batch1"))
b1t<- table(merged.Batch1$SCT_snn_res.1, merged.Batch1$individual)
remove("merged.Batch1")
b1tcolsums<- colSums(b1t)
percb1t<- b1t/b1tcolsums
merged.Batch2<- (subset(merged, Batch == "Batch2"))
b2t<- table(merged.Batch2$SCT_snn_res.1, merged.Batch2$individual)
remove("merged.Batch2")
b2tcolsums<- colSums(b2t)
percb2t<- b2t/b2tcolsums
merged.Batch3<- (subset(merged, Batch == "Batch3"))
b3t<- table(merged.Batch3$SCT_snn_res.1, merged.Batch3$individual)
remove("merged.Batch3")
b3tcolsums<- colSums(b3t)
percb3t<- b3t/b3tcolsumscols1<- c("Batch1_18511","Batch1_18858","Batch1_19160", "Batch2_18511", "Batch2_18858","Batch2_19160",
"Batch3_18511","Batch3_18858", "Batch3_19160")
cols2<- c("Batch1_18511", "Batch2_18511", "Batch3_18511","Batch1_18858", "Batch2_18858", "Batch3_18858","Batch1_19160", "Batch2_19160", "Batch3_19160")
fullpercs<- as.data.frame(cbind(percb1t[,1:3], percb2t,percb3t))
colnames(fullpercs)<-cols1
fullpercs<- cbind(fullpercs$Batch1_18511, fullpercs$Batch2_18511, fullpercs$Batch3_18511,
fullpercs$Batch1_18858, fullpercs$Batch2_18858, fullpercs$Batch3_18858,
fullpercs$Batch1_19160, fullpercs$Batch2_19160, fullpercs$Batch3_19160)
colnames(fullpercs)<-cols2
fullpercs_cor<- round(cor(fullpercs),2)
fullpercs_melt<- melt(fullpercs_cor)
ggplot(data= fullpercs_melt, aes(x=Var1, y=Var2, fill=value)) +
geom_tile(color="white") +
scale_fill_gradient2(low="blue", high="red", mid="white", midpoint = 0, limit= c(-1,1), space= "Lab", name="Pearson\nCorrelation") +
theme_minimal() +
theme(axis.text.x = element_text(angle = 30, hjust=1))+
ggtitle("Pairwise Pearson Correlation of the percent of cells from \neach cell line assigned to each Seurat Cluster (res 1)")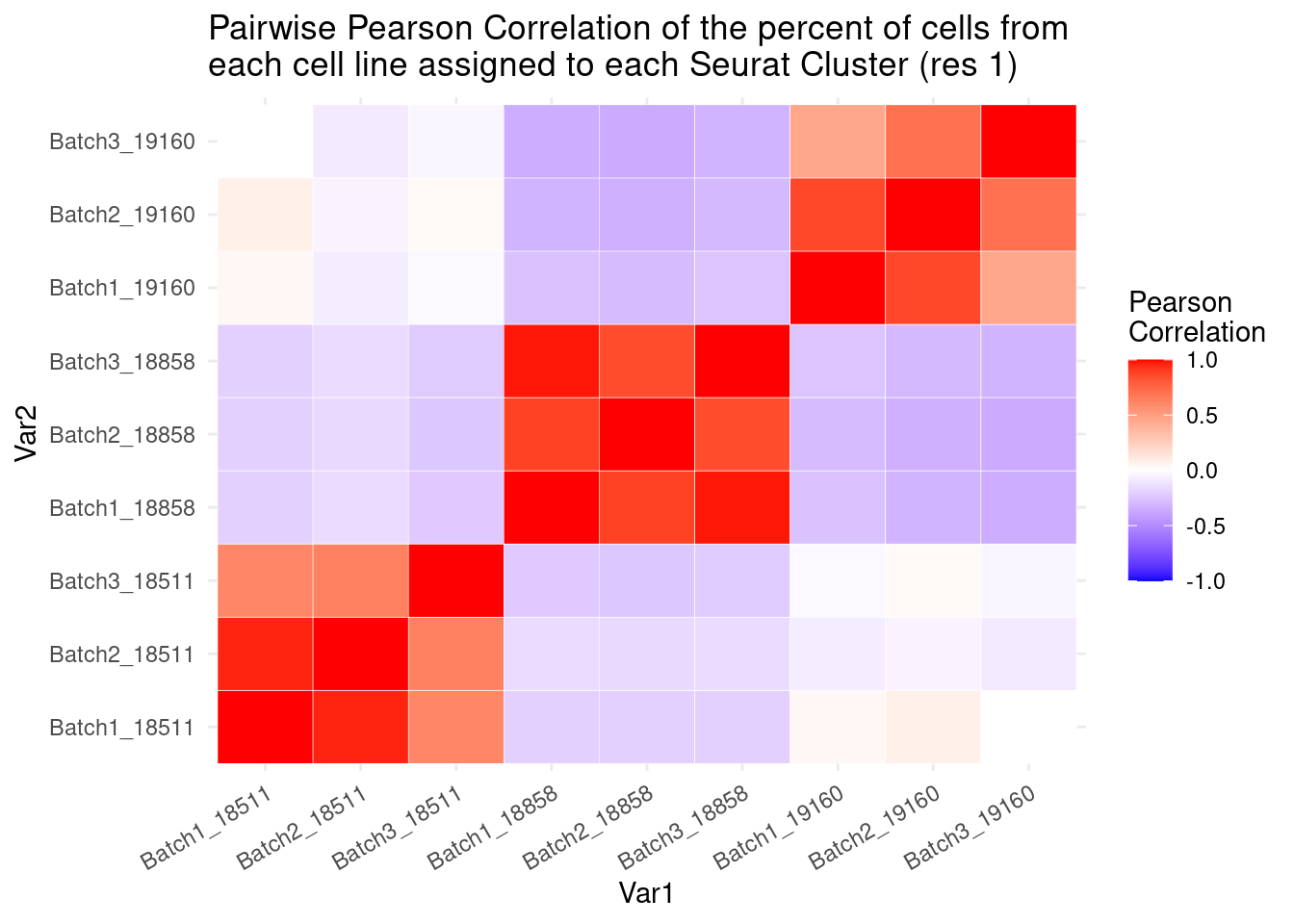
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
#now clustering individual_Batch samples with hierarchical clustering/they will get reordered based on similarity
beauty<- colorRampPalette(brewer.pal(9,"Purples"))(200)
rownames(fullpercs)<- c(0:(nrow(fullpercs)-1))
heatmap(as.matrix(fullpercs), scale="none", col=beauty, cexCol = .7, cexRow=.6)
text(1:ncol(fullpercs),labels=names(fullpercs),srt=30)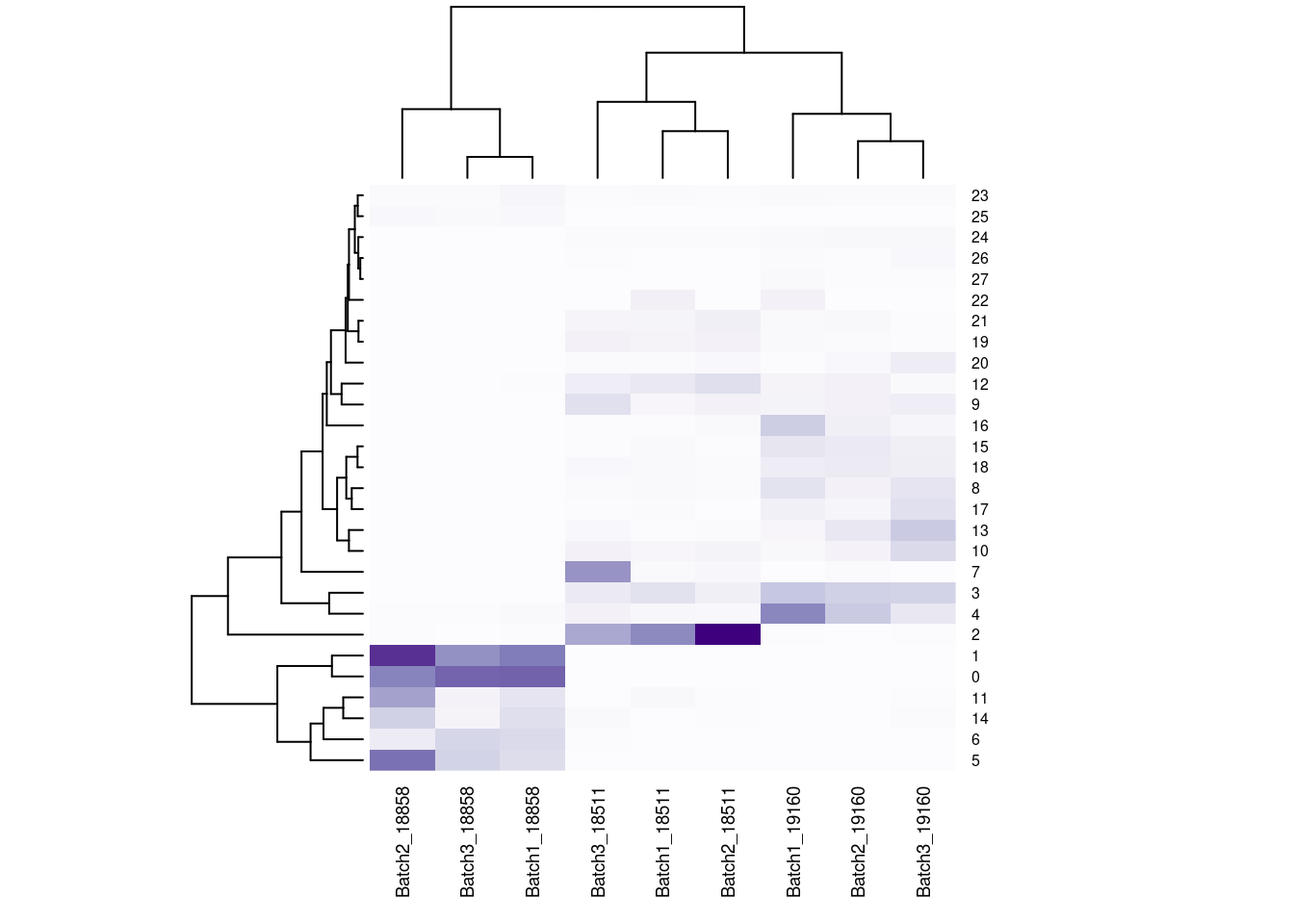
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
#generate a heatmap of the proportion of cells from each individual_batch in each seurat cluster. dendrograms based on similarity of the vectors. should be colored by the value(proportion), but some of the cluster/sample values to seem to match with the colorReclustering with less resolution, check if everything is robust
#reassign idents
Idents(merged)<- 'SCT_snn_res.0.5'DimPlot(merged, reduction = "umap")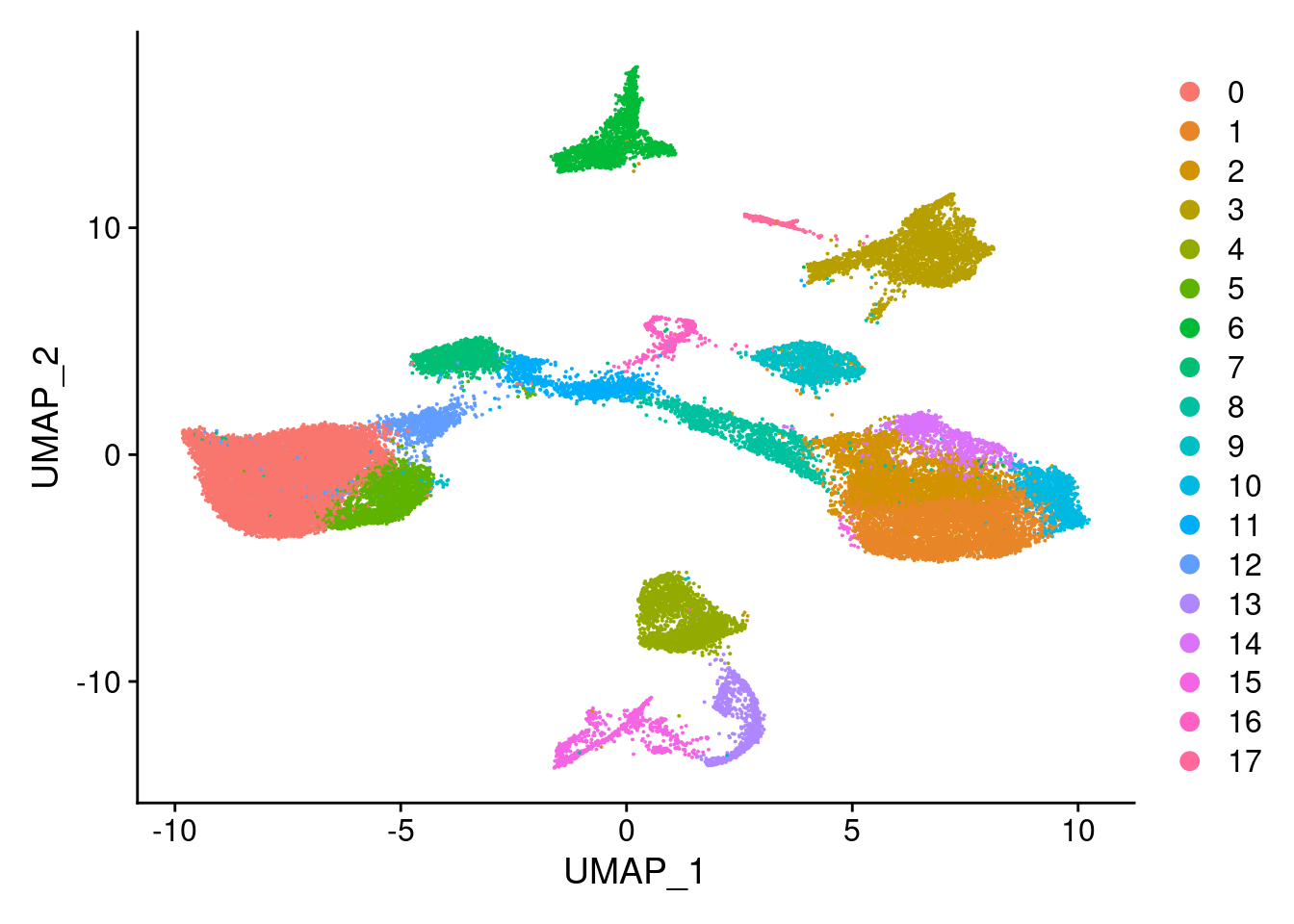
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
DimPlot(merged, reduction = "umap", group.by = "Batch")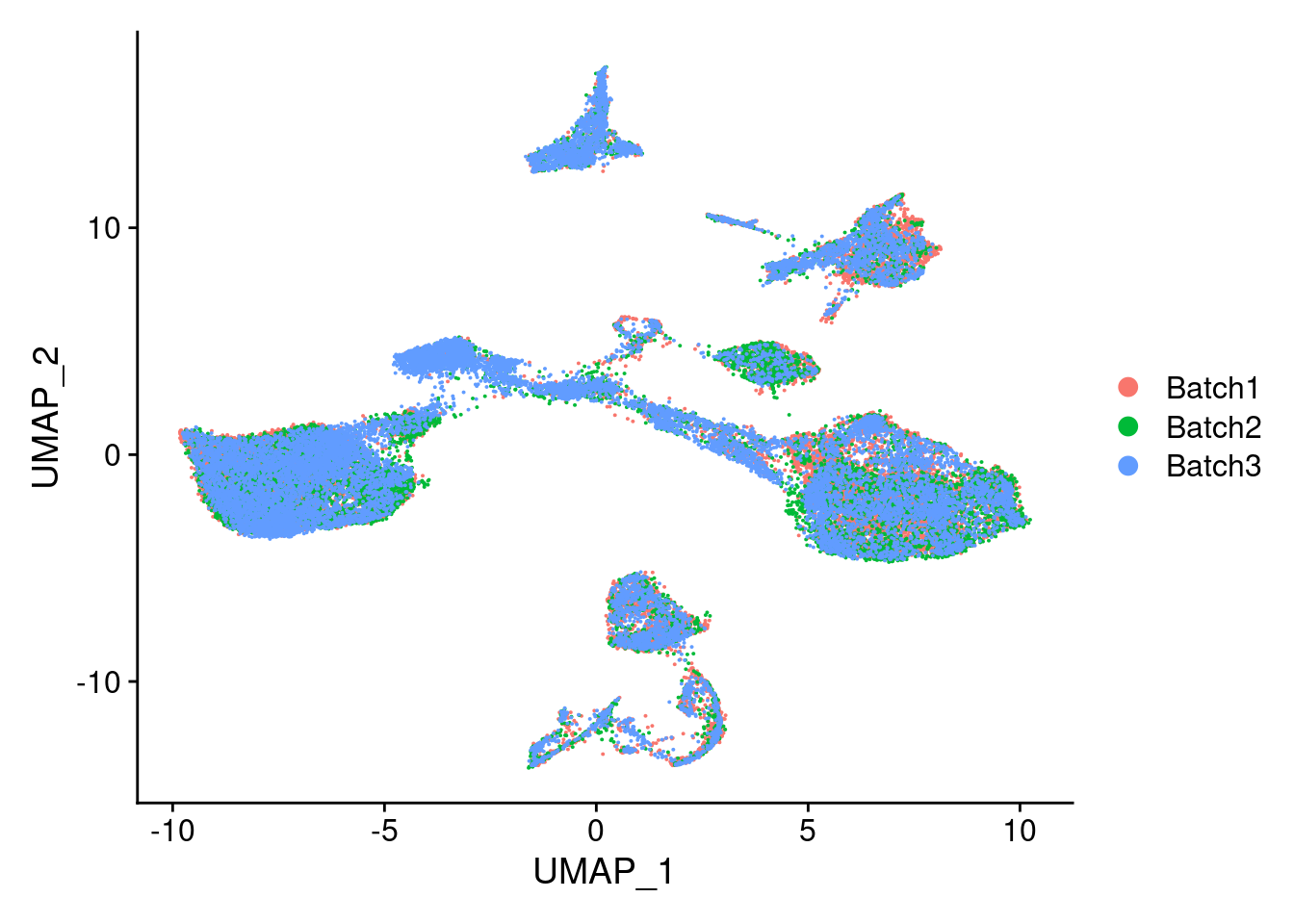
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
DimPlot(merged, reduction = "umap", group.by = "individual")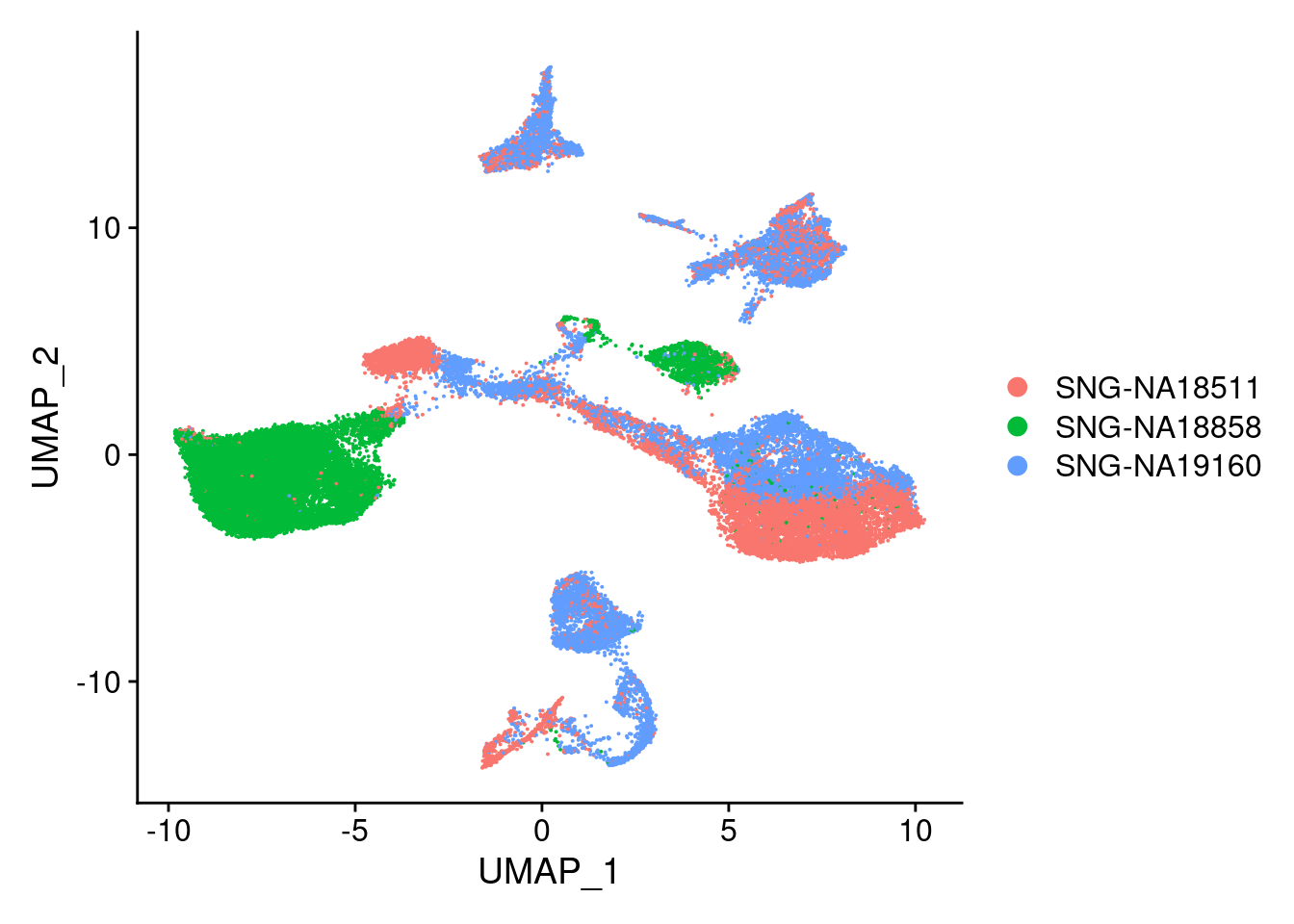
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
xlim <- c(min(merged@reductions$umap@cell.embeddings[,'UMAP_1']),
max(merged@reductions$umap@cell.embeddings[,'UMAP_1']))
ylim <- c(min(merged@reductions$umap@cell.embeddings[,'UMAP_2']),
max(merged@reductions$umap@cell.embeddings[,'UMAP_2']))
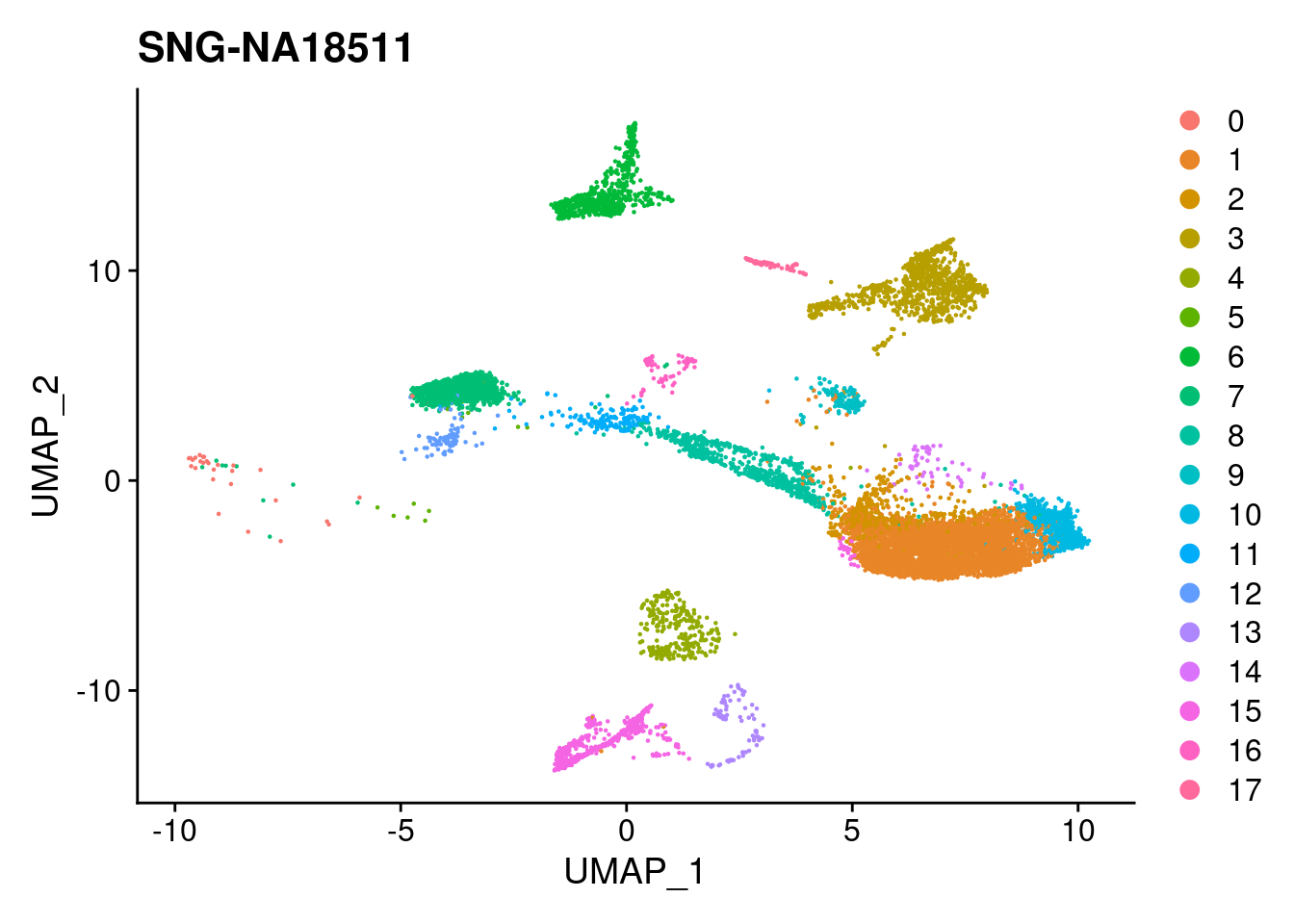
for (i in individuals)
{
print(DimPlot(merged, reduction = "umap",
cells = WhichCells(merged, expression = individual == i)) +
xlim(xlim) + ylim(ylim) + ggtitle(i))
}
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
#exploring similarity in the number of cells per individual between batches
merged.Batch1<- (subset(merged, Batch == "Batch1"))
b1t<- table(merged.Batch1$SCT_snn_res.0.5, merged.Batch1$individual)
remove("merged.Batch1")
b1tcolsums<- colSums(b1t)
percb1t<- b1t/b1tcolsums
merged.Batch2<- (subset(merged, Batch == "Batch2"))
b2t<- table(merged.Batch2$SCT_snn_res.0.5, merged.Batch2$individual)
remove("merged.Batch2")
b2tcolsums<- colSums(b2t)
percb2t<- b2t/b2tcolsums
merged.Batch3<- (subset(merged, Batch == "Batch3"))
b3t<- table(merged.Batch3$SCT_snn_res.0.5, merged.Batch3$individual)
remove("merged.Batch3")
b3tcolsums<- colSums(b3t)
percb3t<- b3t/b3tcolsumscols1<- c("Batch1_18511","Batch1_18858","Batch1_19160", "Batch2_18511", "Batch2_18858","Batch2_19160",
"Batch3_18511","Batch3_18858", "Batch3_19160")
cols2<- c("Batch1_18511", "Batch2_18511", "Batch3_18511","Batch1_18858", "Batch2_18858", "Batch3_18858","Batch1_19160", "Batch2_19160", "Batch3_19160")
fullpercs<- as.data.frame(cbind(percb1t[,1:3], percb2t,percb3t))
colnames(fullpercs)<-cols1
fullpercs<- cbind(fullpercs$Batch1_18511, fullpercs$Batch2_18511, fullpercs$Batch3_18511,
fullpercs$Batch1_18858, fullpercs$Batch2_18858, fullpercs$Batch3_18858,
fullpercs$Batch1_19160, fullpercs$Batch2_19160, fullpercs$Batch3_19160)
colnames(fullpercs)<-cols2
fullpercs_cor<- round(cor(fullpercs),2)
fullpercs_melt<- melt(fullpercs_cor)
ggplot(data= fullpercs_melt, aes(x=Var1, y=Var2, fill=value)) +
geom_tile(color="white") +
scale_fill_gradient2(low="blue", high="red", mid="white", midpoint = 0, limit= c(-1,1), space= "Lab", name="Pearson\nCorrelation") +
theme_minimal() +
theme(axis.text.x = element_text(angle = 30, hjust=1))+
ggtitle("Pairwise Pearson Correlation of the percent of cells from \neach cell line assigned to each Seurat Cluster\ncluster res. 0.5")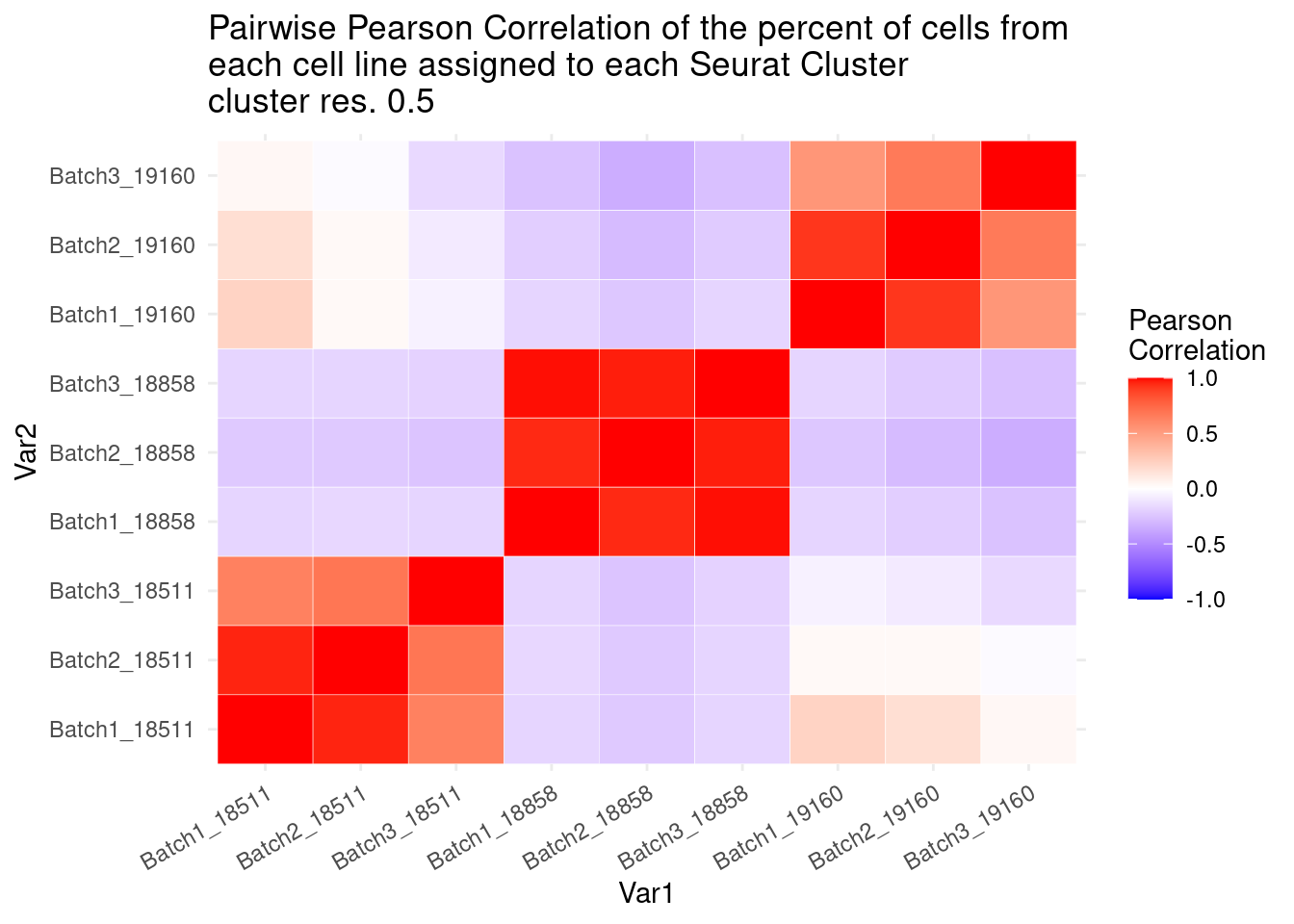
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
#now clustering individual_Batch samples with hierarchical clustering/they will get reordered based on similarity
beauty<- colorRampPalette(brewer.pal(9,"Purples"))(200)
rownames(fullpercs)<- c(0:(nrow(fullpercs)-1))
heatmap(as.matrix(fullpercs), scale="none", col=beauty, cexCol = .7, cexRow=.6)
text(1:ncol(fullpercs),labels=names(fullpercs),srt=30)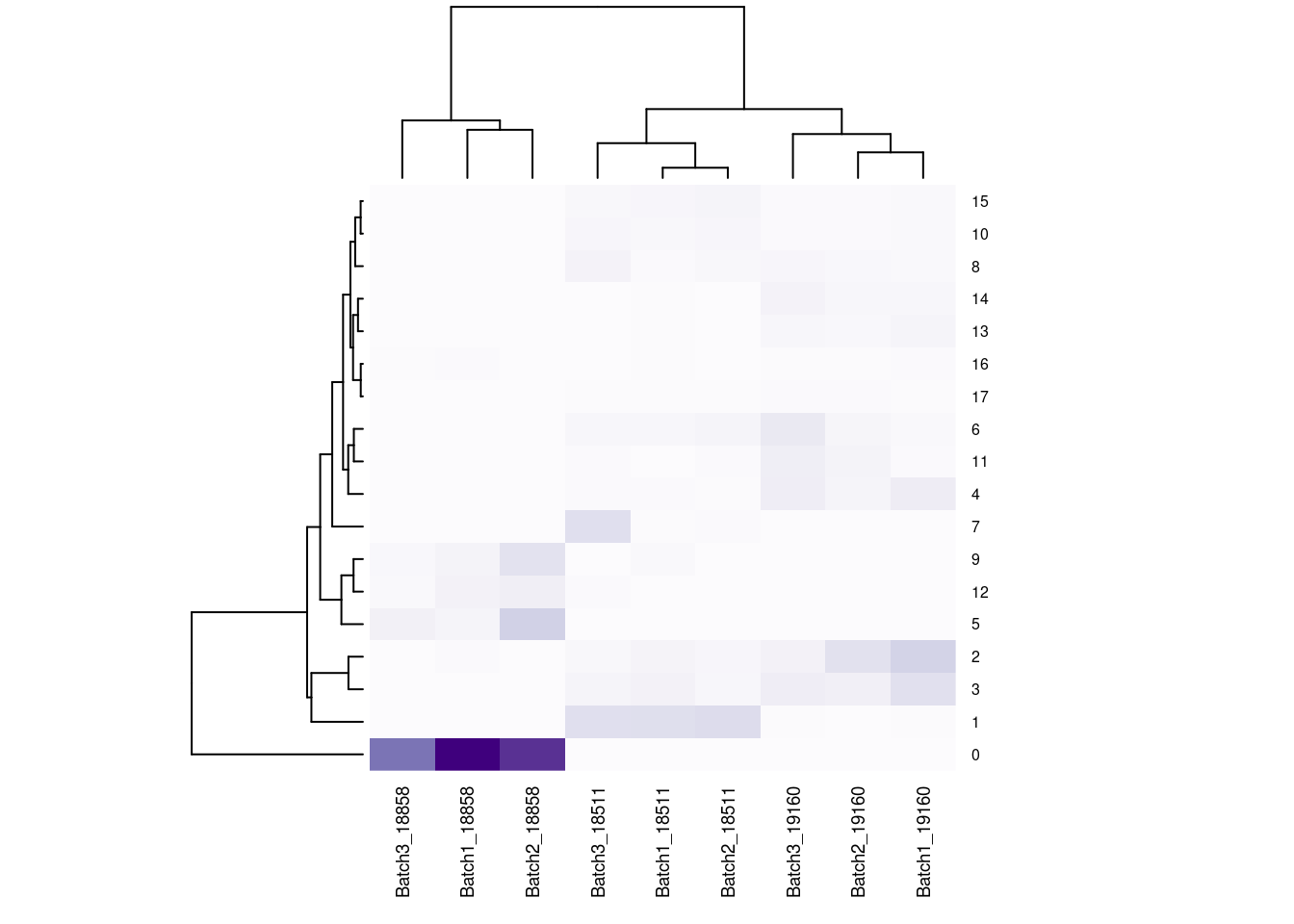
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
#generate a heatmap of the raw proportion of cells from each individual_batch in each seurat cluster. dendrograms based on similarity of the vectors. should be colored by the value(proportion), but some of the cluster/sample values to seem to match with the color#reassign idents
Idents(merged)<- 'SCT_snn_res.0.1'DimPlot(merged, reduction = "umap")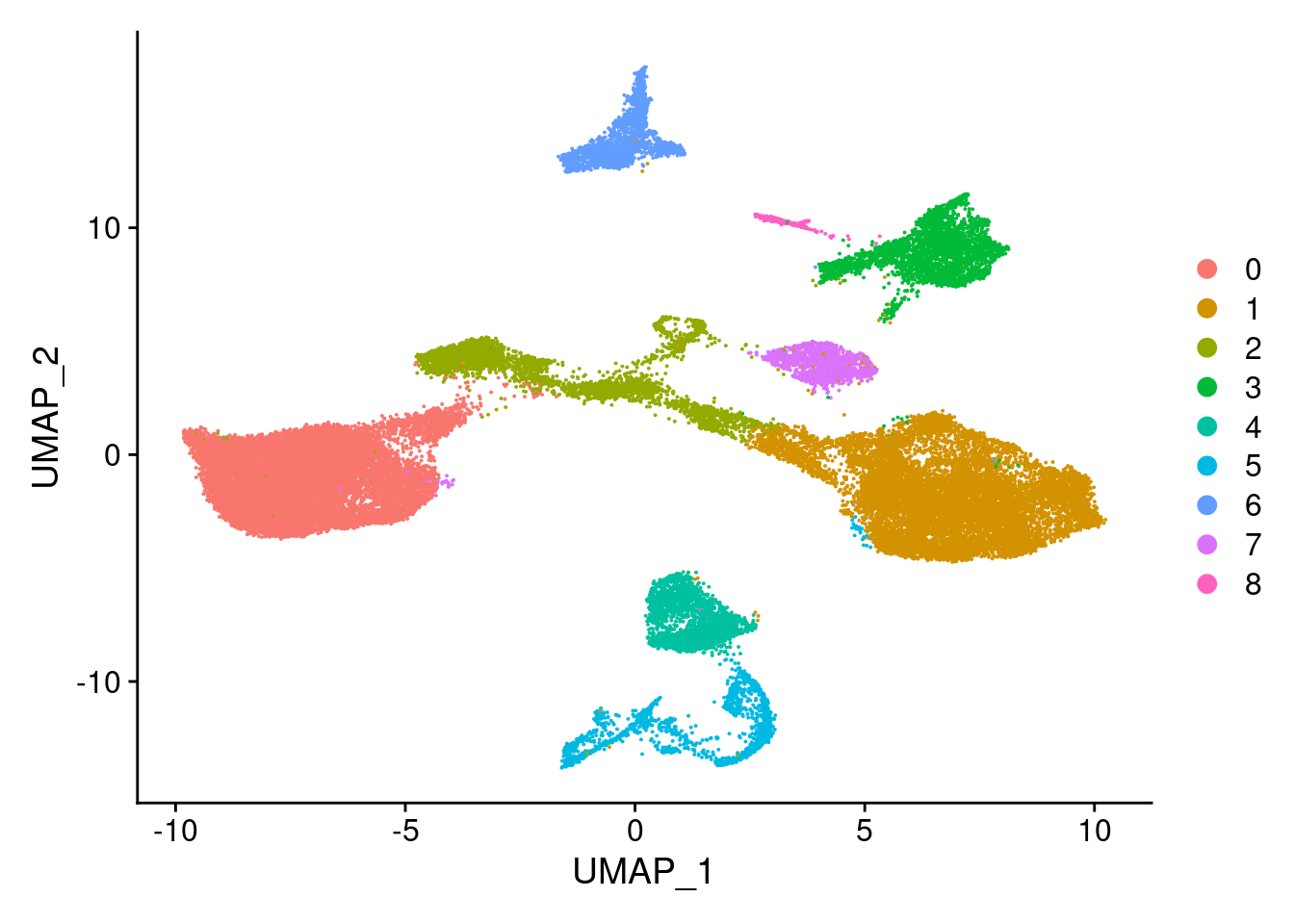
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
DimPlot(merged, reduction = "umap", group.by = "Batch")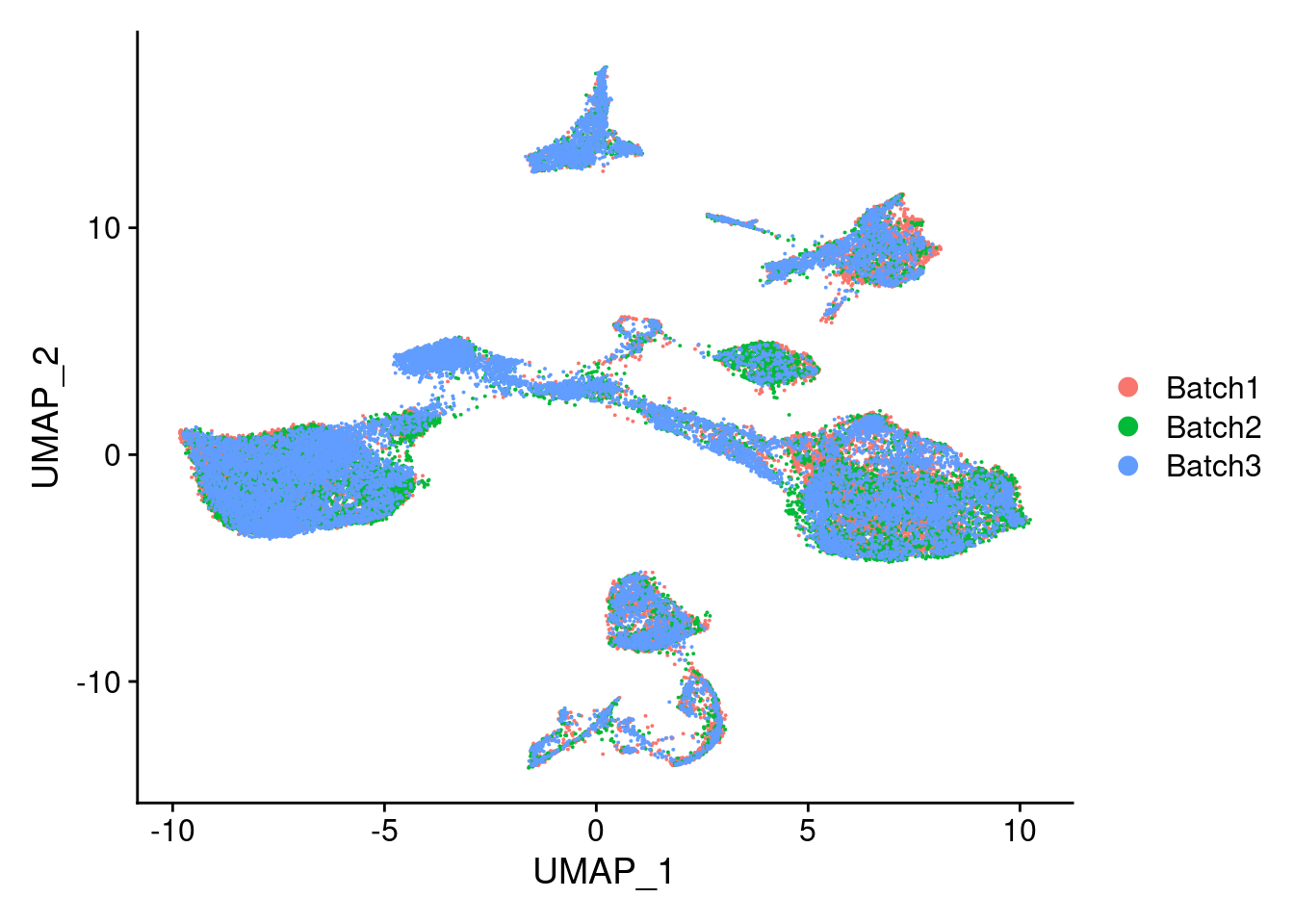
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
DimPlot(merged, reduction = "umap", group.by = "individual")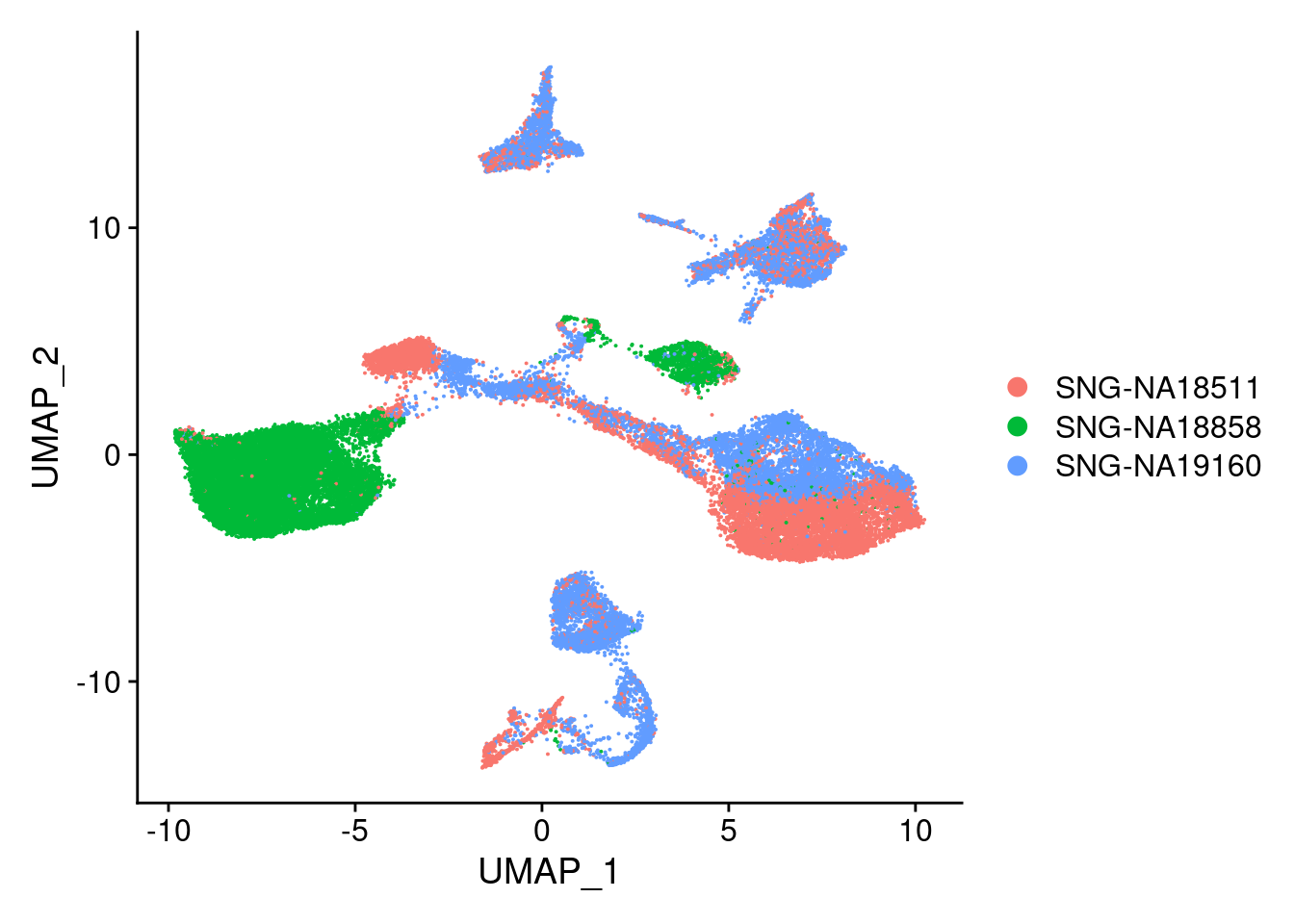
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
xlim <- c(min(merged@reductions$umap@cell.embeddings[,'UMAP_1']),
max(merged@reductions$umap@cell.embeddings[,'UMAP_1']))
ylim <- c(min(merged@reductions$umap@cell.embeddings[,'UMAP_2']),
max(merged@reductions$umap@cell.embeddings[,'UMAP_2']))
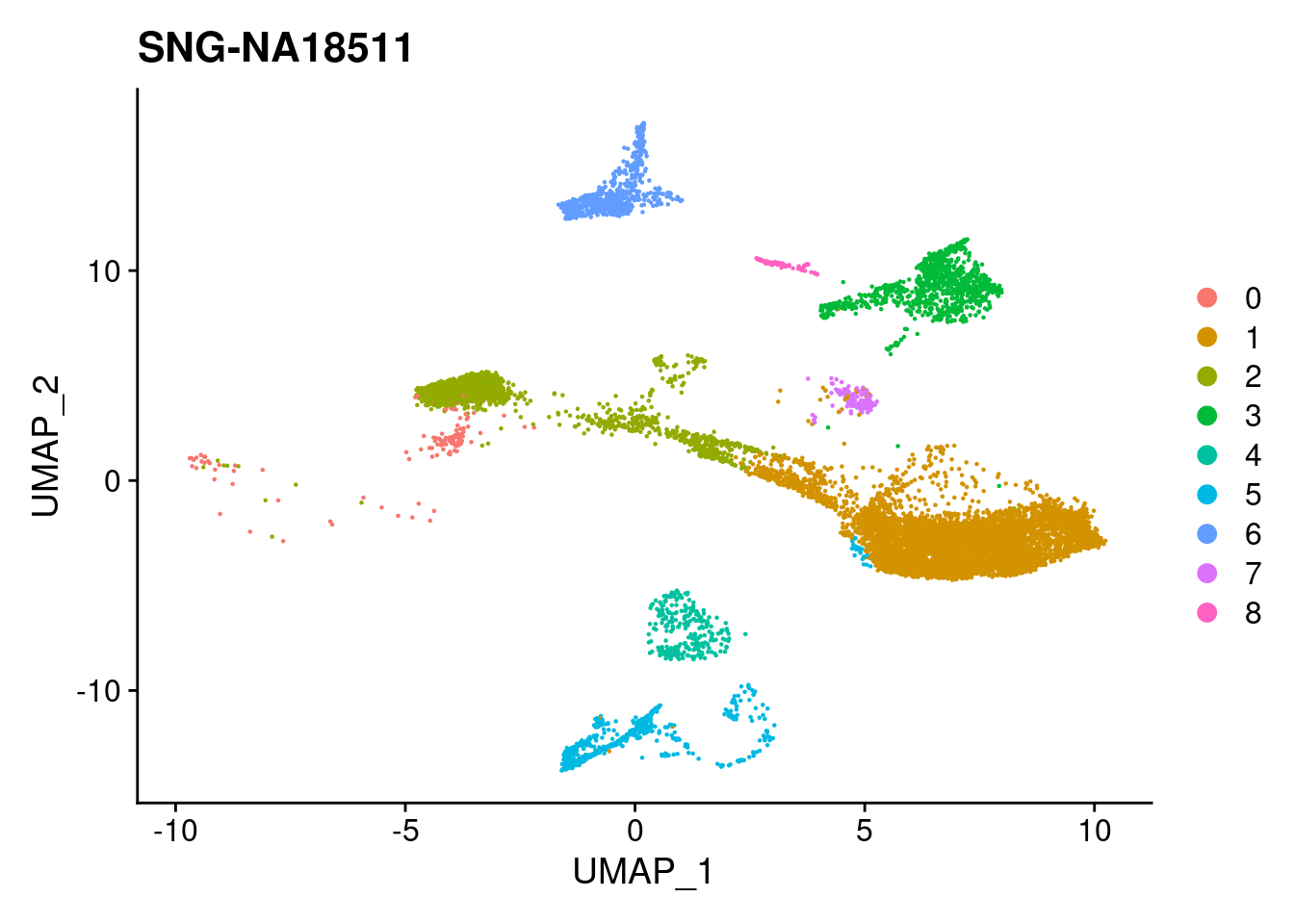
for (i in individuals)
{
print(DimPlot(merged, reduction = "umap",
cells = WhichCells(merged, expression = individual == i)) +
xlim(xlim) + ylim(ylim) + ggtitle(i))
}
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
#exploring similarity in the number of cells per individual between batches
merged.Batch1<- (subset(merged, Batch == "Batch1"))
b1t<- table(merged.Batch1$SCT_snn_res.0.1, merged.Batch1$individual)
remove("merged.Batch1")
b1tcolsums<- colSums(b1t)
percb1t<- b1t/b1tcolsums
merged.Batch2<- (subset(merged, Batch == "Batch2"))
b2t<- table(merged.Batch2$SCT_snn_res.0.1, merged.Batch2$individual)
remove("merged.Batch2")
b2tcolsums<- colSums(b2t)
percb2t<- b2t/b2tcolsums
merged.Batch3<- (subset(merged, Batch == "Batch3"))
b3t<- table(merged.Batch3$SCT_snn_res.0.1, merged.Batch3$individual)
remove("merged.Batch3")
b3tcolsums<- colSums(b3t)
percb3t<- b3t/b3tcolsumscols1<- c("Batch1_18511","Batch1_18858","Batch1_19160", "Batch2_18511", "Batch2_18858","Batch2_19160",
"Batch3_18511","Batch3_18858", "Batch3_19160")
cols2<- c("Batch1_18511", "Batch2_18511", "Batch3_18511","Batch1_18858", "Batch2_18858", "Batch3_18858","Batch1_19160", "Batch2_19160", "Batch3_19160")
fullpercs<- as.data.frame(cbind(percb1t[,1:3], percb2t,percb3t))
colnames(fullpercs)<-cols1
fullpercs<- cbind(fullpercs$Batch1_18511, fullpercs$Batch2_18511, fullpercs$Batch3_18511,
fullpercs$Batch1_18858, fullpercs$Batch2_18858, fullpercs$Batch3_18858,
fullpercs$Batch1_19160, fullpercs$Batch2_19160, fullpercs$Batch3_19160)
colnames(fullpercs)<-cols2
fullpercs_cor<- round(cor(fullpercs),2)
fullpercs_melt<- melt(fullpercs_cor)
ggplot(data= fullpercs_melt, aes(x=Var1, y=Var2, fill=value)) +
geom_tile(color="white") +
scale_fill_gradient2(low="blue", high="red", mid="white", midpoint = 0, limit= c(-1,1), space= "Lab", name="Pearson\nCorrelation") +
theme_minimal() +
theme(axis.text.x = element_text(angle = 30, hjust=1))+
ggtitle("Pairwise Pearson Correlation of the percent of cells from \neach cell line assigned to each Seurat Cluster\ncluster res. 0.1")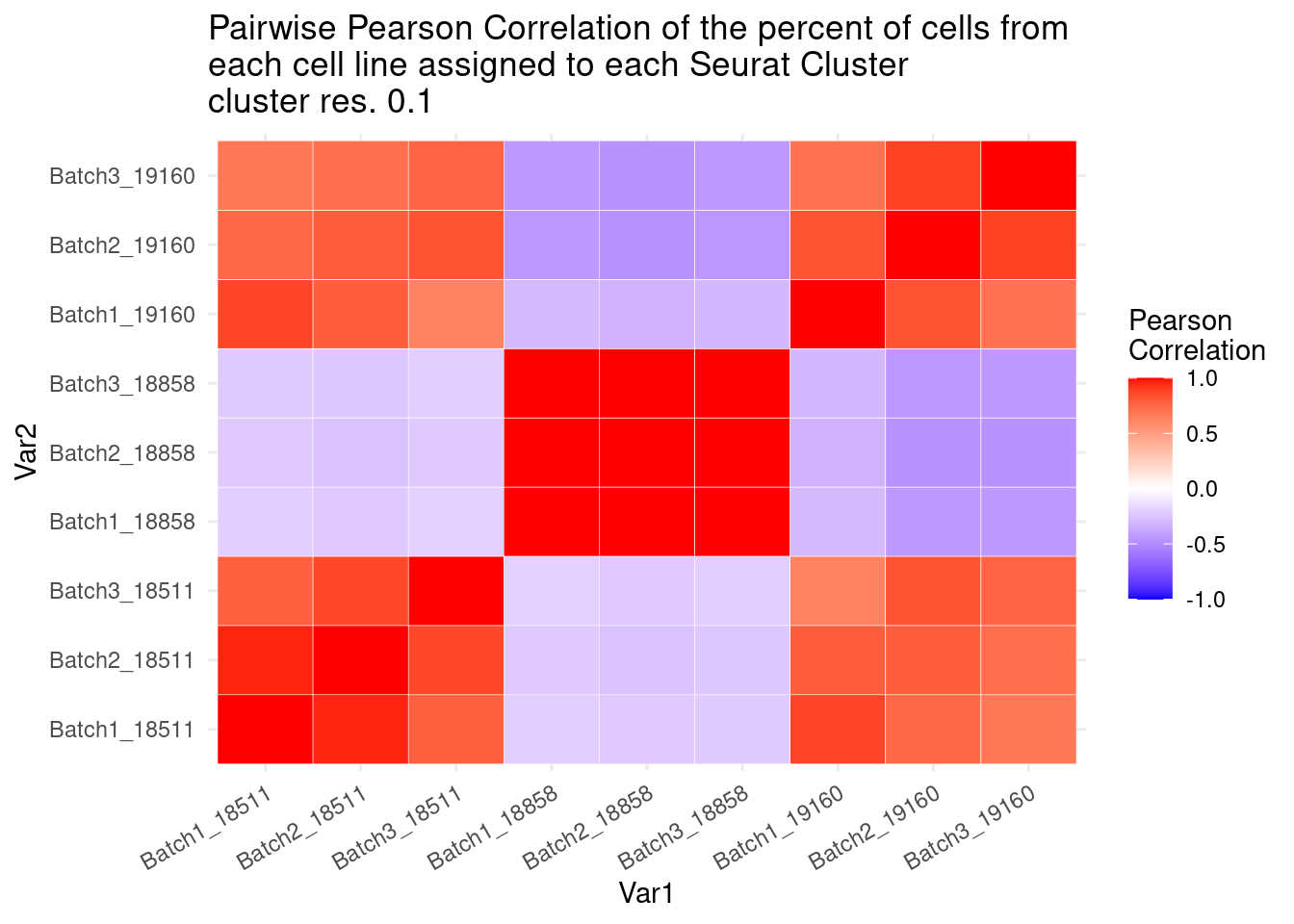
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
#now clustering individual_Batch samples with hierarchical clustering/they will get reordered based on similarity
beauty<- colorRampPalette(brewer.pal(9,"Purples"))(200)
#fullnonorm<- as.data.frame(cbind(b1t[,1:3], b2t,b3t))
#colnames(fullnonorm)<-cols1
#heatmap((as.matrix(fullnonorm)), scale="column", col= beauty)
rownames(fullpercs)<- c(0:(nrow(fullpercs)-1))
heatmap(as.matrix(fullpercs), scale="none", col=beauty, cexCol = .7, cexRow=.6)
text(1:ncol(fullpercs),labels=names(fullpercs),srt=30)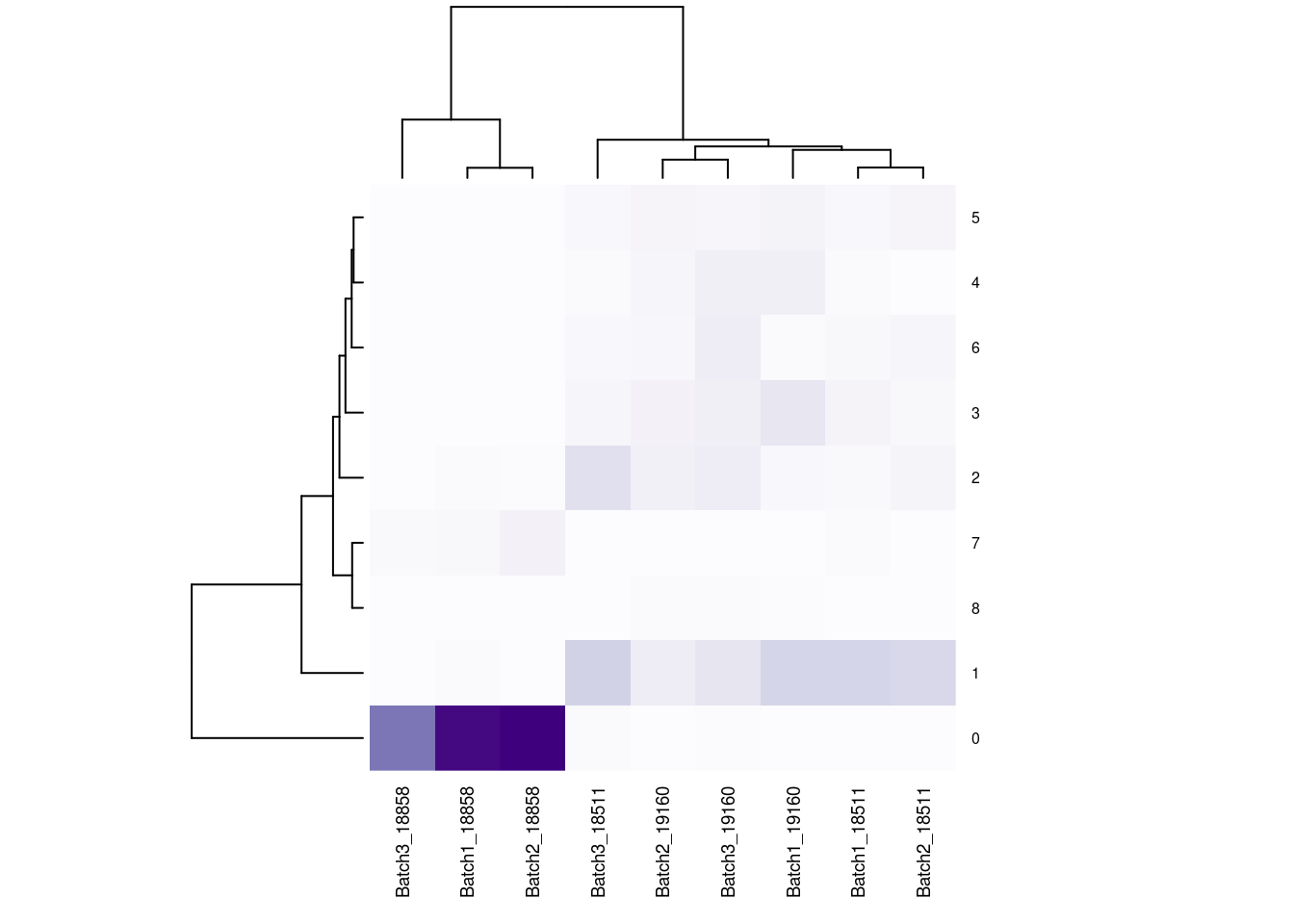
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
#generate a heatmap of the raw proportion of cells from each individual_batch in each seurat cluster. dendrograms based on similarity of the vectors. should be colored by the value(proportion), but some of the cluster/sample values to seem to match with the color#reassign idents
Idents(merged)<- 'SCT_snn_res.0.8'DimPlot(merged, reduction = "umap")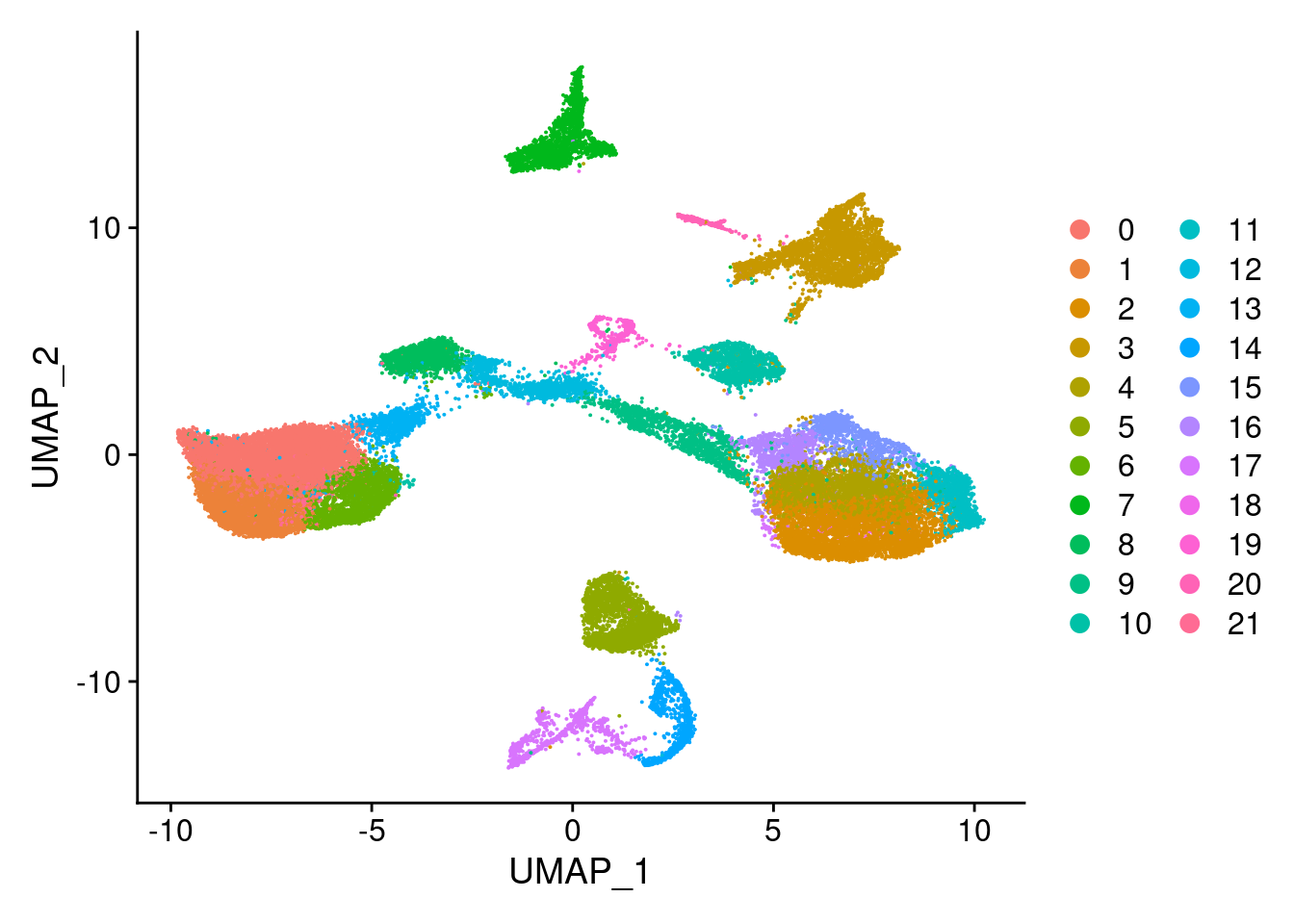
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
DimPlot(merged, reduction = "umap", group.by = "Batch")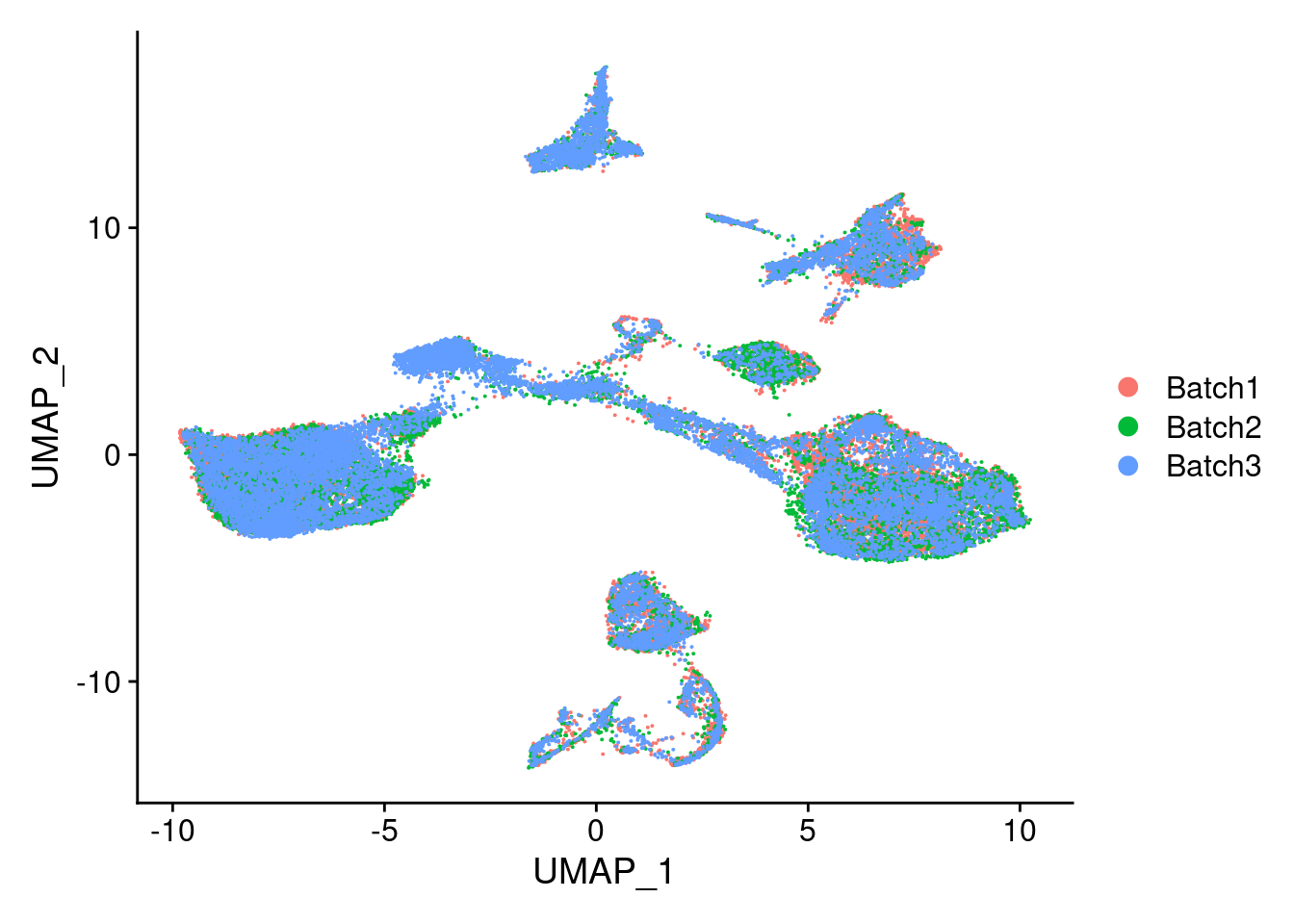
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
DimPlot(merged, reduction = "umap", group.by = "individual")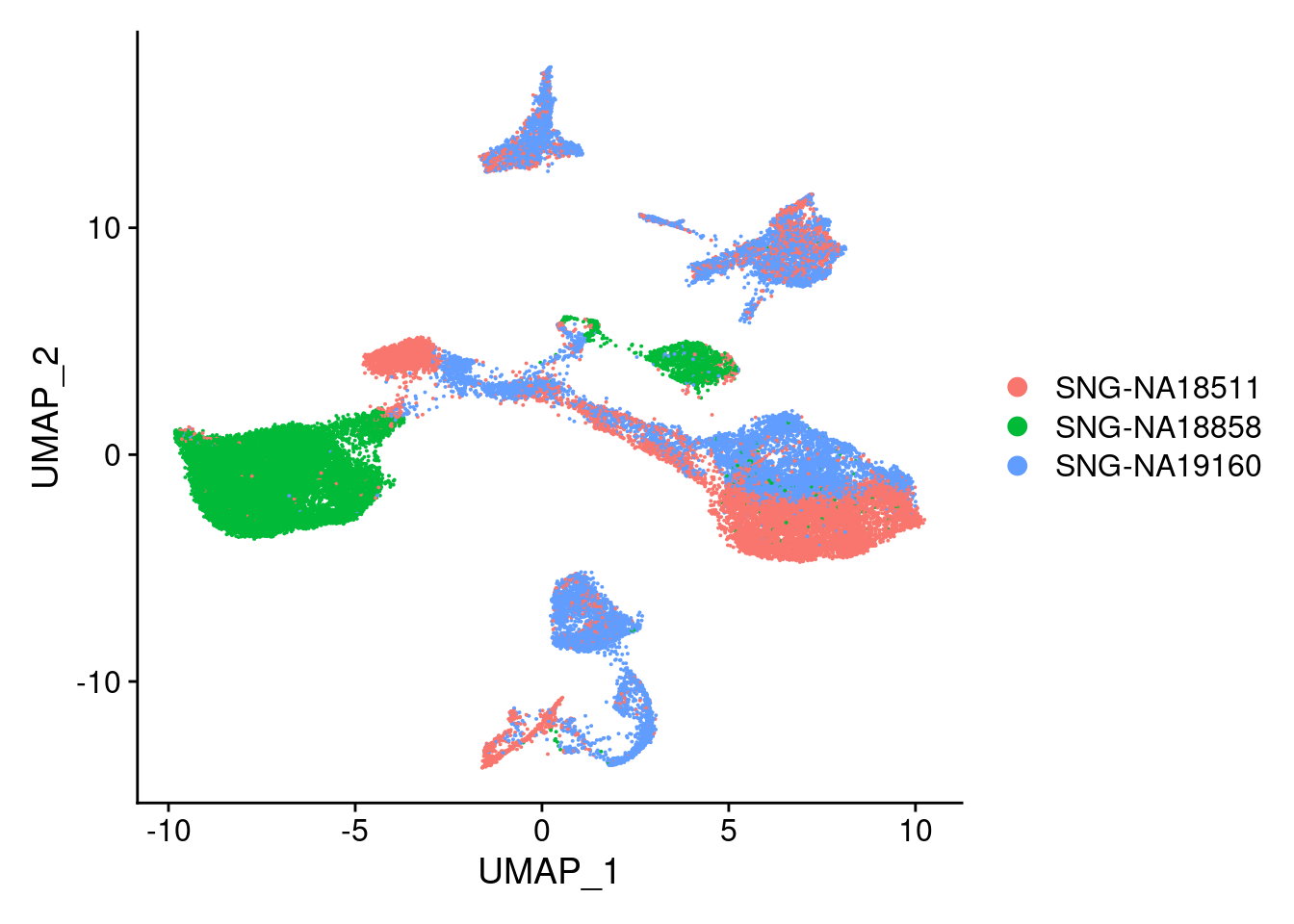
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
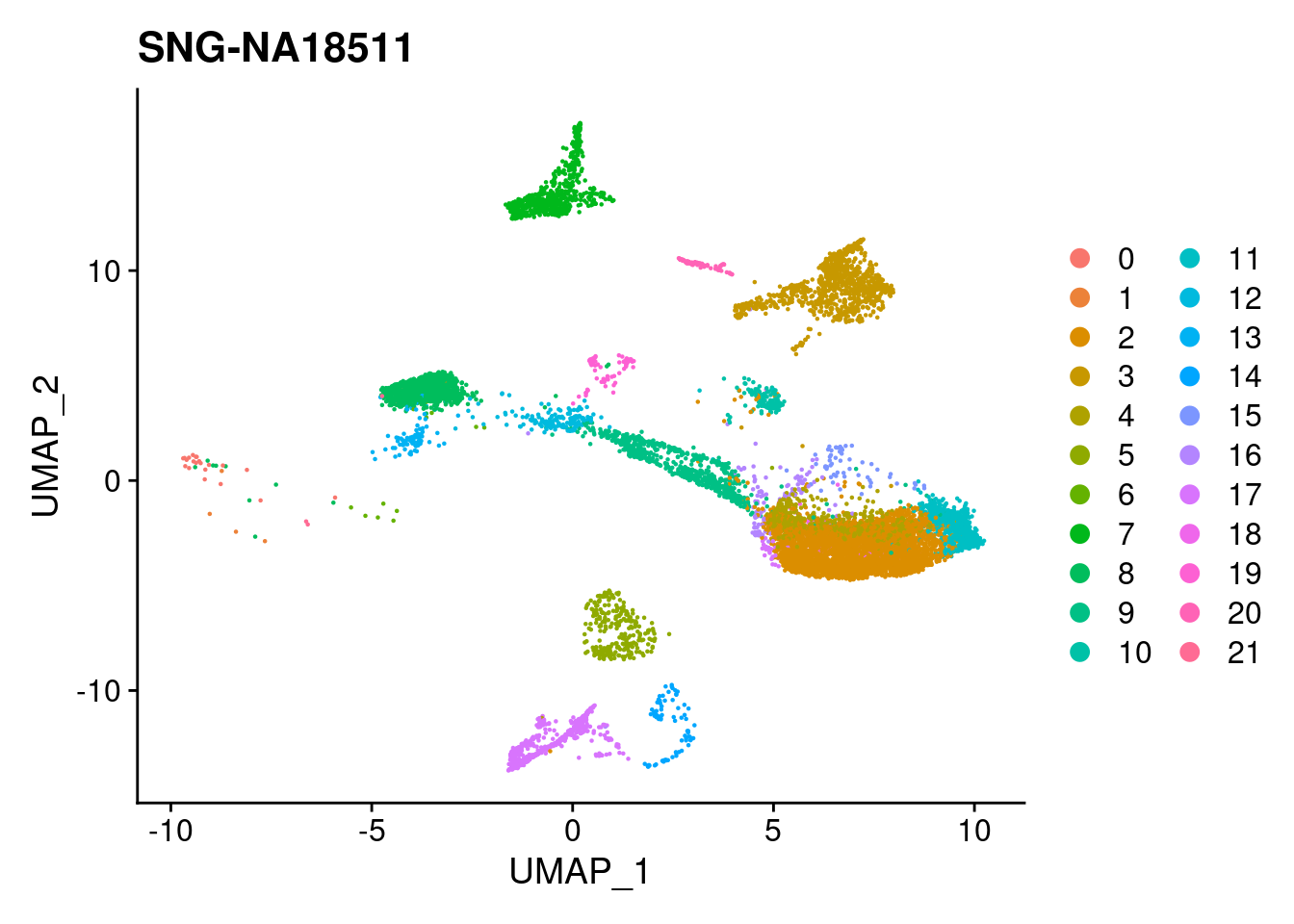
xlim <- c(min(merged@reductions$umap@cell.embeddings[,'UMAP_1']),
max(merged@reductions$umap@cell.embeddings[,'UMAP_1']))
ylim <- c(min(merged@reductions$umap@cell.embeddings[,'UMAP_2']),
max(merged@reductions$umap@cell.embeddings[,'UMAP_2']))
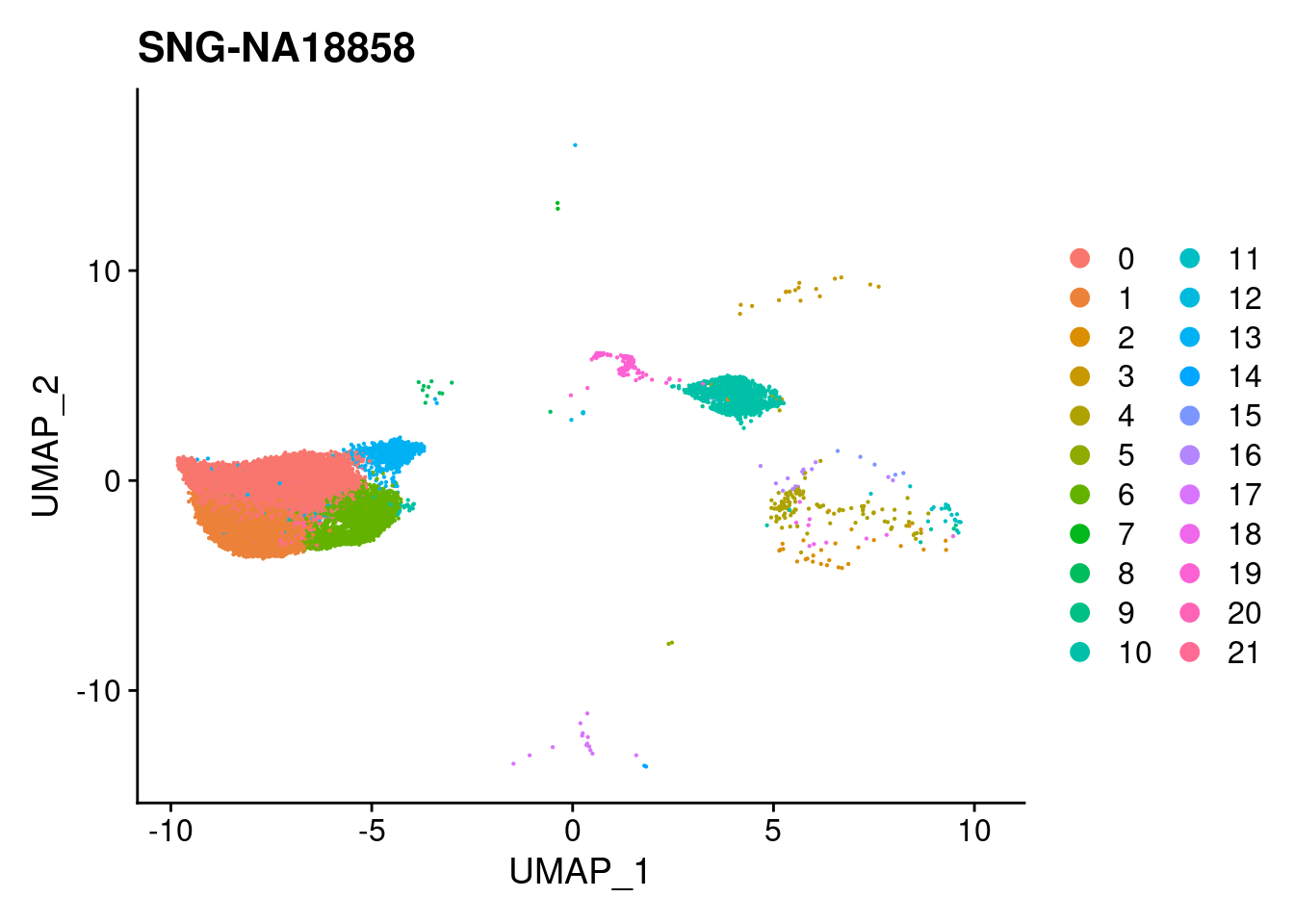
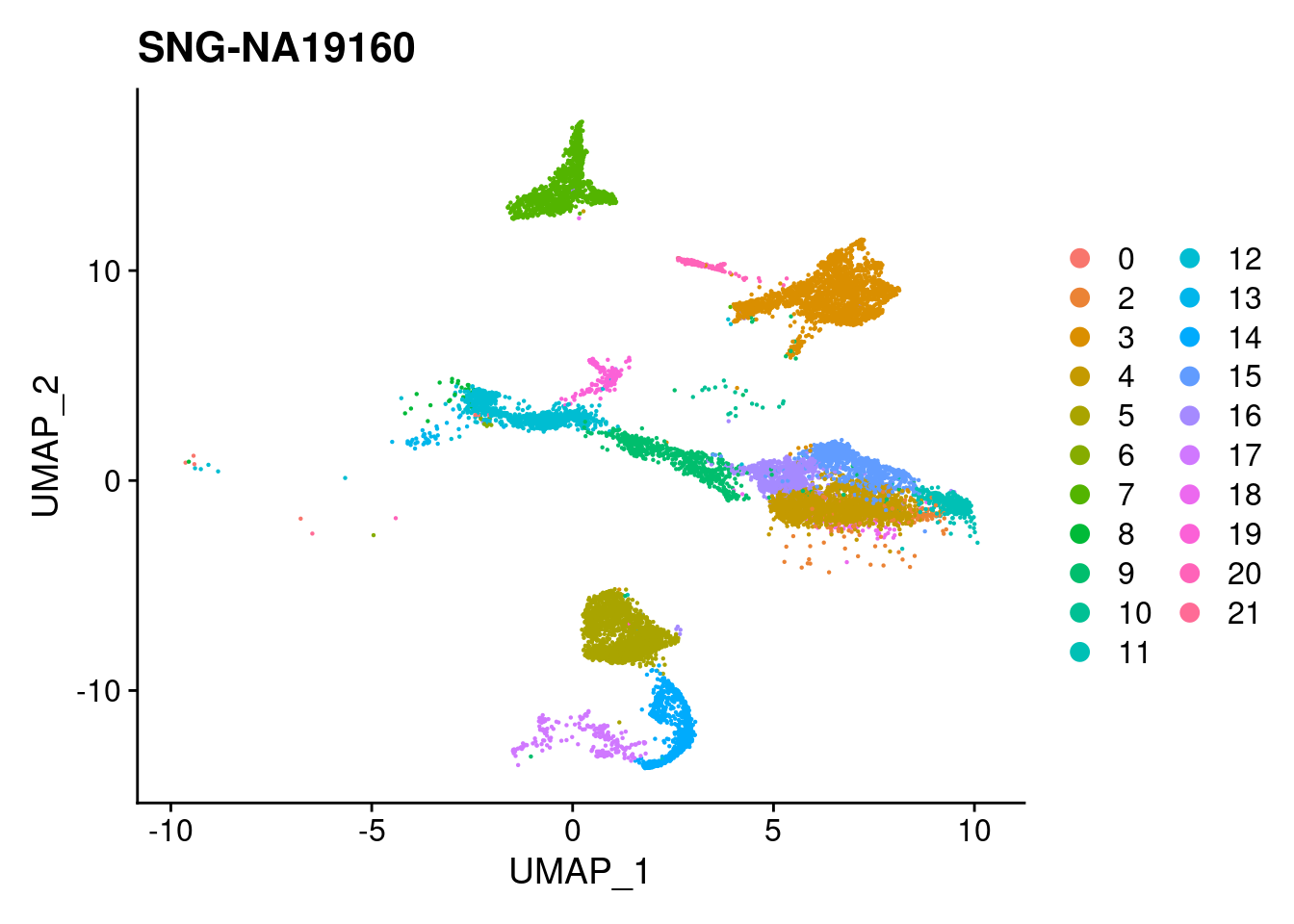
for (i in individuals)
{
print(DimPlot(merged, reduction = "umap",
cells = WhichCells(merged, expression = individual == i)) +
xlim(xlim) + ylim(ylim) + ggtitle(i))
}
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
#exploring similarity in the number of cells per individual between batches
merged.Batch1<- (subset(merged, Batch == "Batch1"))
b1t<- table(merged.Batch1$SCT_snn_res.0.8, merged.Batch1$individual)
remove("merged.Batch1")
b1tcolsums<- colSums(b1t)
percb1t<- b1t/b1tcolsums
merged.Batch2<- (subset(merged, Batch == "Batch2"))
b2t<- table(merged.Batch2$SCT_snn_res.0.8, merged.Batch2$individual)
remove("merged.Batch2")
b2tcolsums<- colSums(b2t)
percb2t<- b2t/b2tcolsums
merged.Batch3<- (subset(merged, Batch == "Batch3"))
b3t<- table(merged.Batch3$SCT_snn_res.0.8, merged.Batch3$individual)
remove("merged.Batch3")
b3tcolsums<- colSums(b3t)
percb3t<- b3t/b3tcolsumscols1<- c("Batch1_18511","Batch1_18858","Batch1_19160", "Batch2_18511", "Batch2_18858","Batch2_19160",
"Batch3_18511","Batch3_18858", "Batch3_19160")
cols2<- c("Batch1_18511", "Batch2_18511", "Batch3_18511","Batch1_18858", "Batch2_18858", "Batch3_18858","Batch1_19160", "Batch2_19160", "Batch3_19160")
fullpercs<- as.data.frame(cbind(percb1t[,1:3], percb2t,percb3t))
colnames(fullpercs)<-cols1
fullpercs<- cbind(fullpercs$Batch1_18511, fullpercs$Batch2_18511, fullpercs$Batch3_18511,
fullpercs$Batch1_18858, fullpercs$Batch2_18858, fullpercs$Batch3_18858,
fullpercs$Batch1_19160, fullpercs$Batch2_19160, fullpercs$Batch3_19160)
colnames(fullpercs)<-cols2
fullpercs_cor<- round(cor(fullpercs),2)
fullpercs_melt<- melt(fullpercs_cor)
ggplot(data= fullpercs_melt, aes(x=Var1, y=Var2, fill=value)) +
geom_tile(color="white") +
scale_fill_gradient2(low="blue", high="red", mid="white", midpoint = 0, limit= c(-1,1), space= "Lab", name="Pearson\nCorrelation") +
theme_minimal() +
ggtitle("Pairwise Pearson Correlation of the percent of cells from \neach cell line assigned to each Seurat Cluster\ncluster res. 0.8")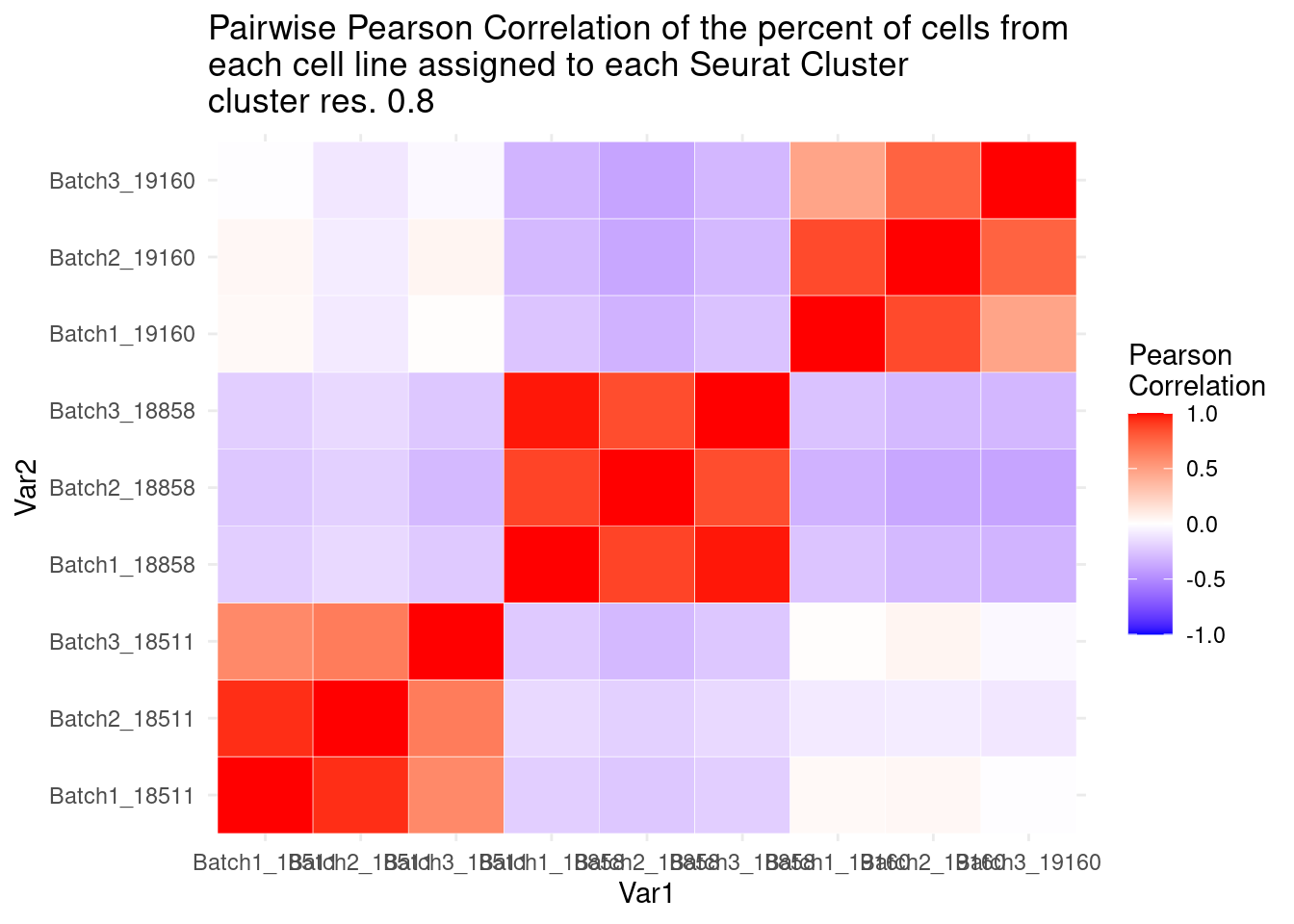
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
#now clustering individual_Batch samples with hierarchical clustering/they will get reordered based on similarity
beauty<- colorRampPalette(brewer.pal(9,"Purples"))(200)
rownames(fullpercs)<- c(0:(nrow(fullpercs)-1))
heatmap(as.matrix(fullpercs), scale="none", col=beauty, cexCol = .7, cexRow=.6)
text(1:ncol(fullpercs),labels=names(fullpercs),srt=30)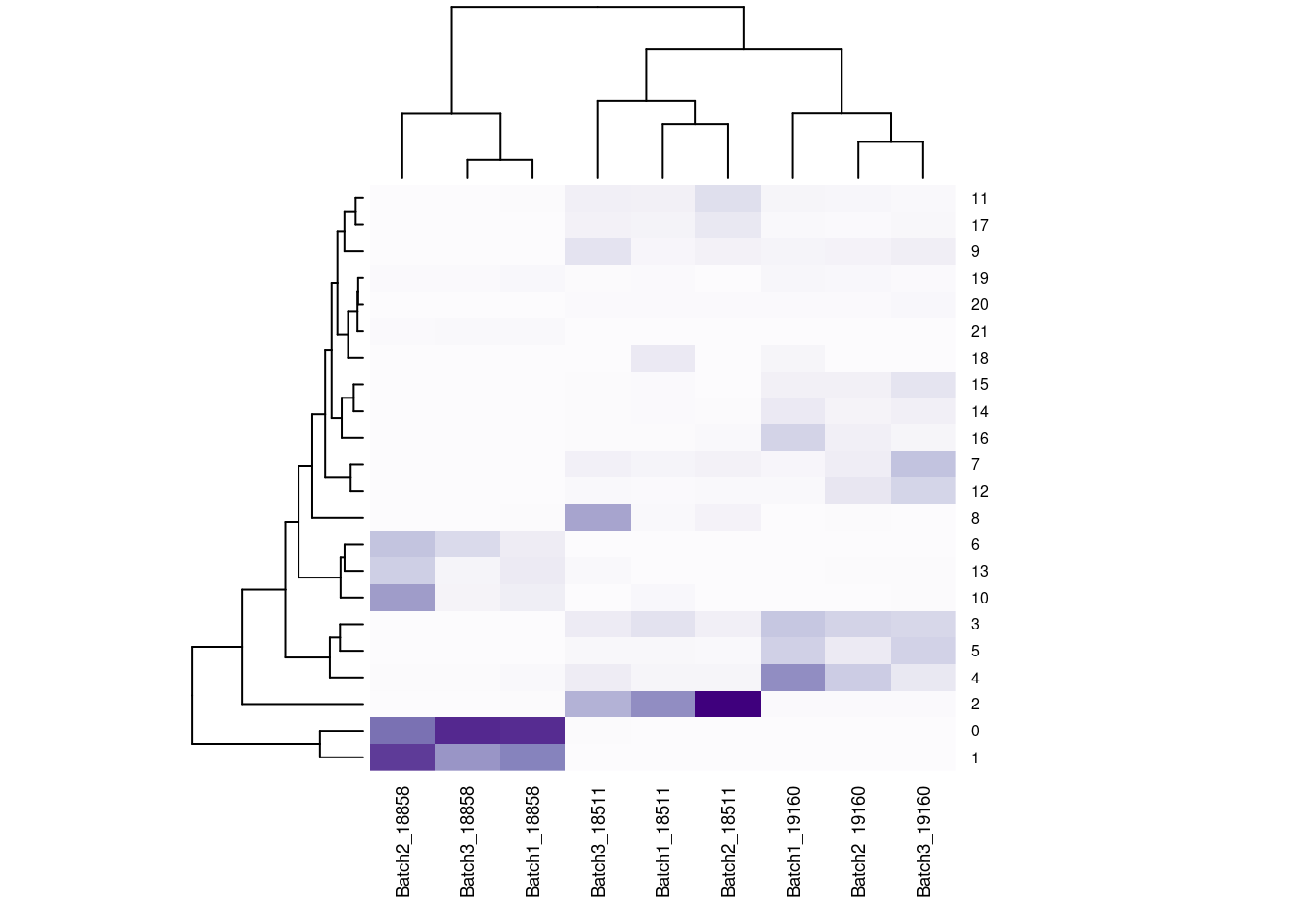
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
#generate a heatmap of the raw proportion of cells from each individual_batch in each seurat cluster. dendrograms based on similarity of the vectors. should be colored by the value(proportion), but some of the cluster/sample values to seem to match with the colorVlnPlot(merged, features= "percent.mt", group.by = "SCT_snn_res.1", pt.size = 0)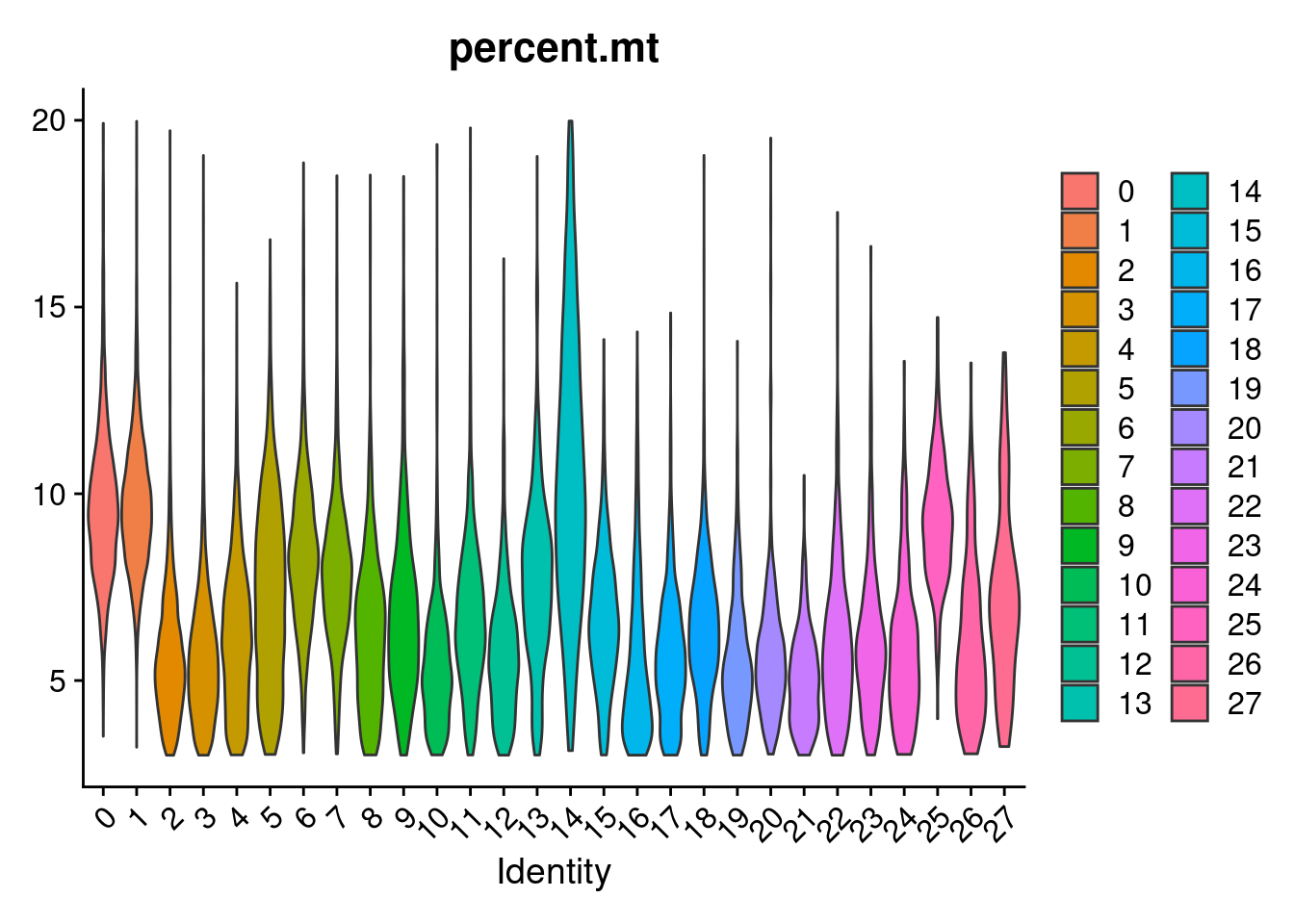
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
merged[["percent.rps"]]<- PercentageFeatureSet(merged, pattern = "^RPS")
merged[["percent.rpl"]]<- PercentageFeatureSet(merged, pattern = "^RPL")
merged[["percent.rp"]]<- merged[["percent.rps"]]+merged[["percent.rpl"]]
VlnPlot(merged, features= "percent.rp", group.by = "SCT_snn_res.1", pt.size=0)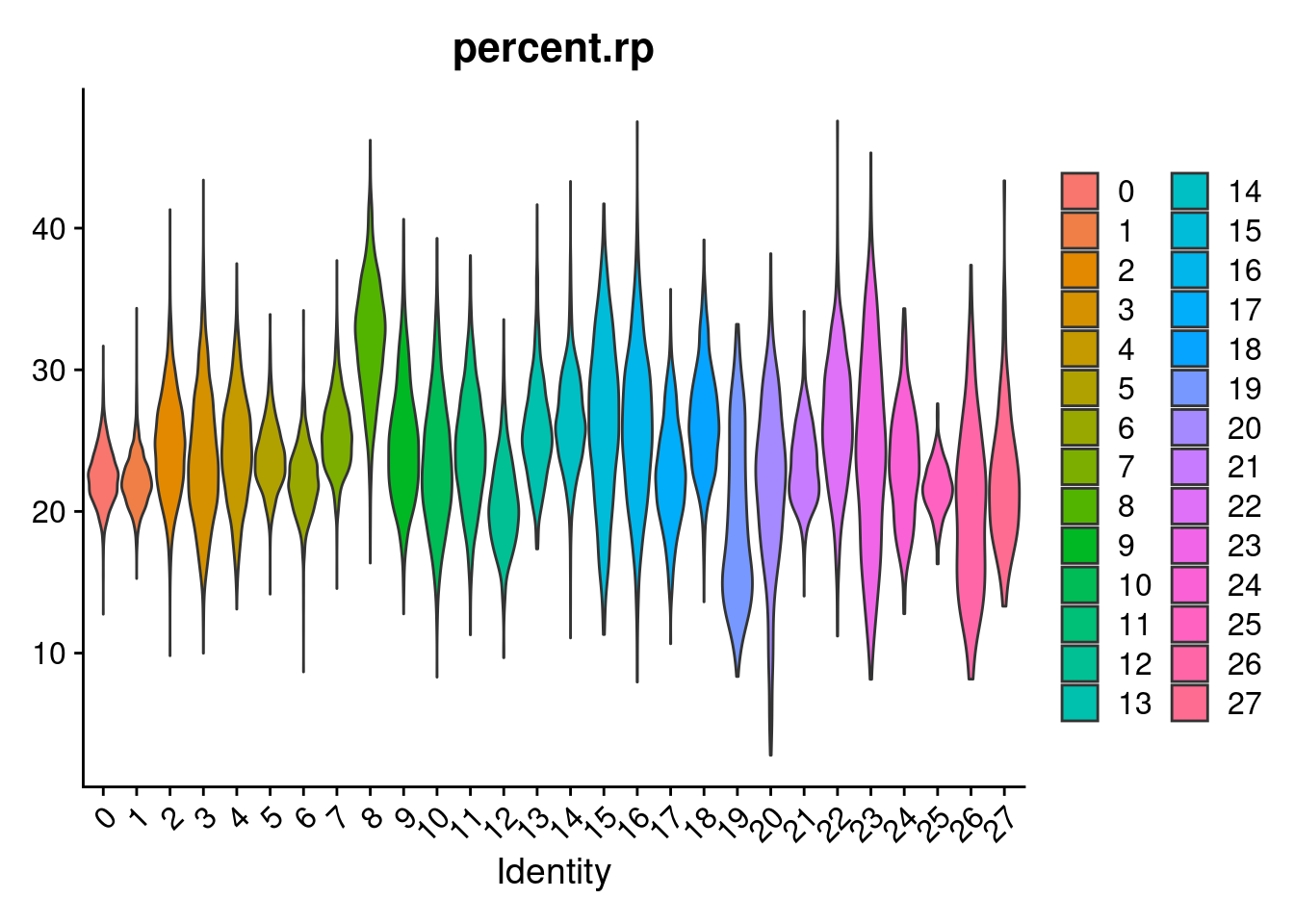
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
FeaturePlot(merged, features = "nFeature_RNA")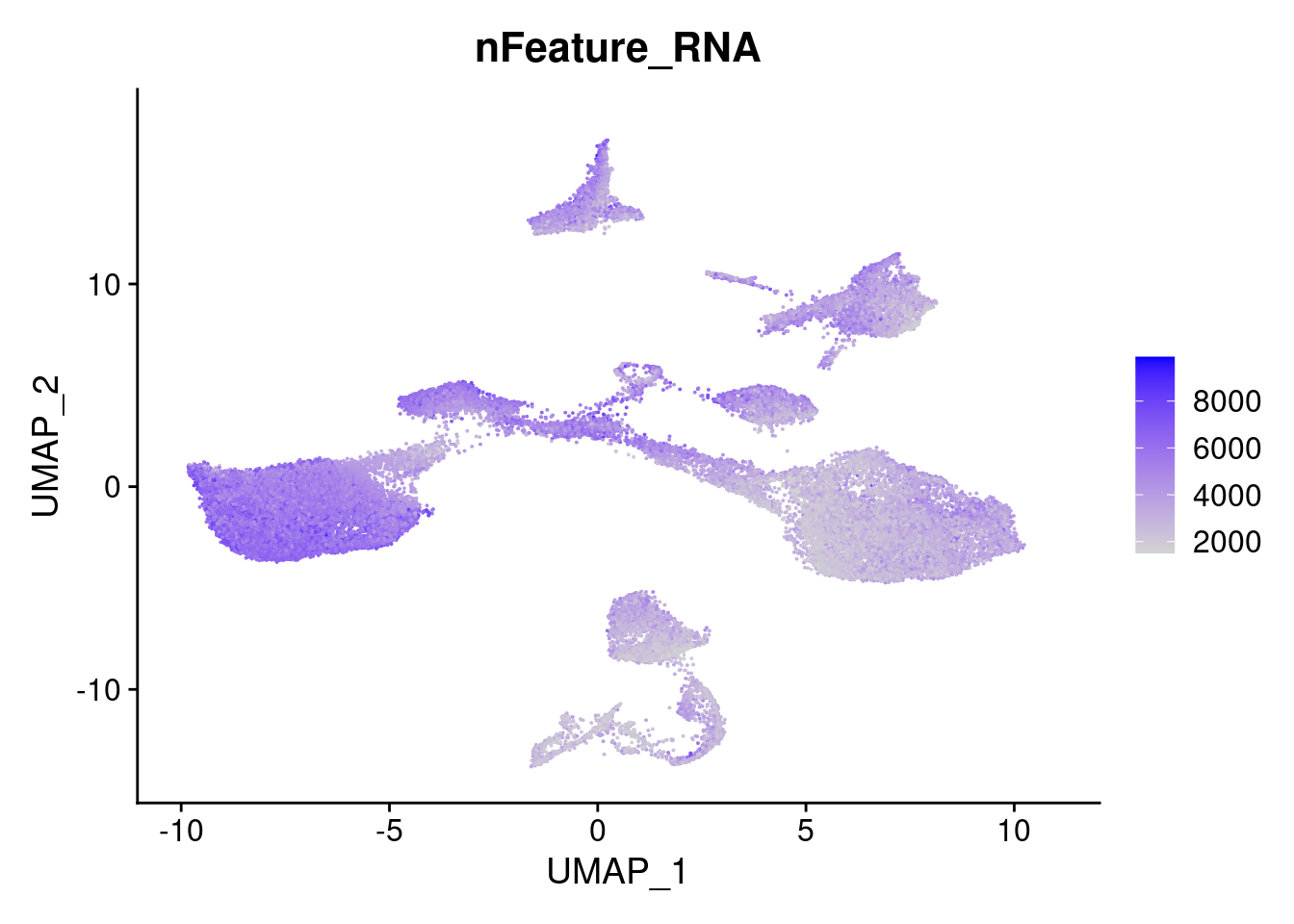
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
head(merged)An object of class Seurat
2 features across 42488 samples within 2 assays
Active assay: SCT (1 features, 1 variable features)
1 other assay present: RNA
3 dimensional reductions calculated: pca, harmony, umapVlnPlot(merged, features= "nFeature_RNA", group.by = "SCT_snn_res.1", pt.size=0)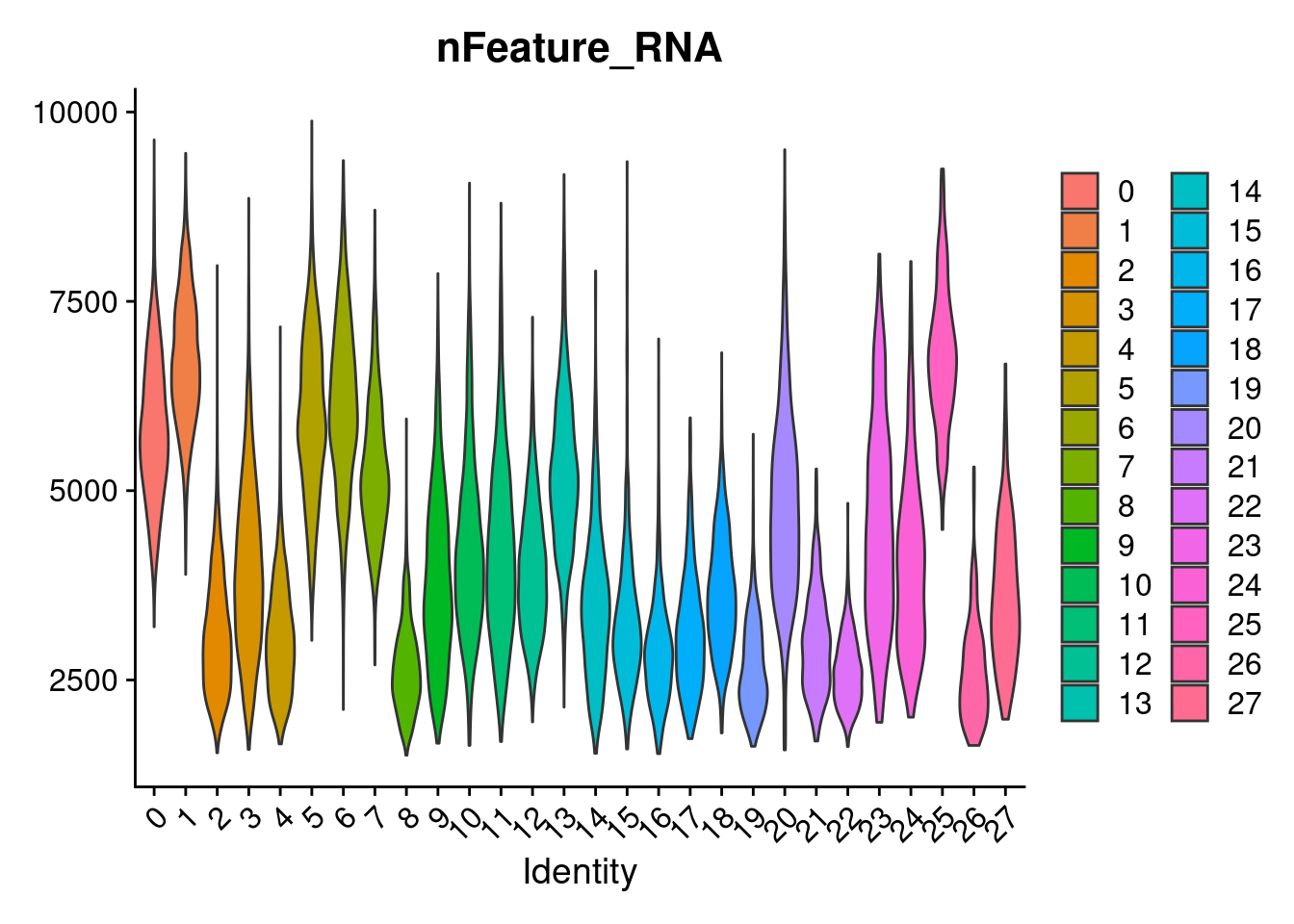
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
FeaturePlot(merged, features = c("POU5F1", "SOX17", "HAND1", "PAX6"), pt.size = 0.2, ncol=2, combine=T)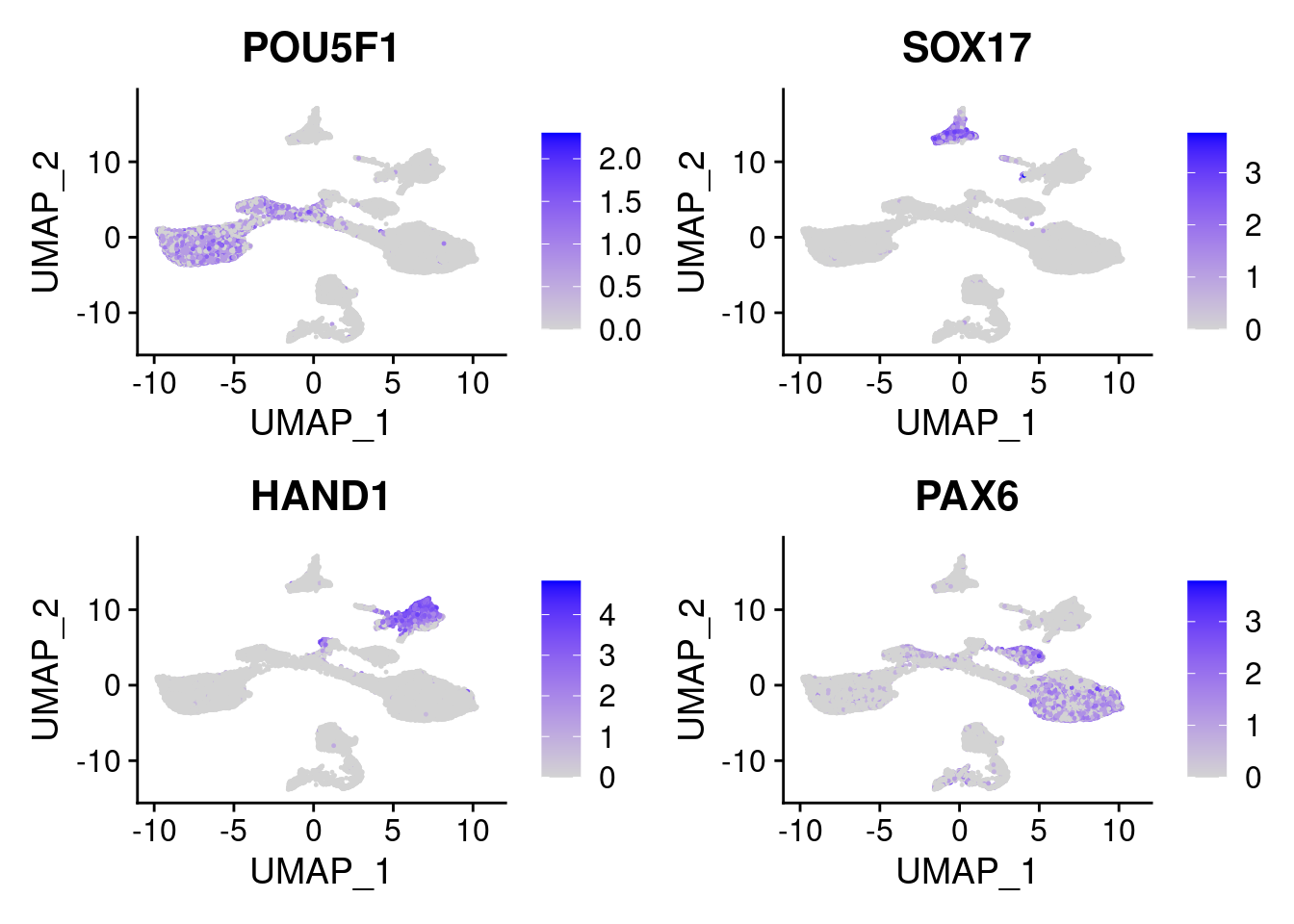
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
FeaturePlot(merged, features = c("FGB", "ECSCR", "NEUROD1", "SOX10"), pt.size = 0.2, ncol=2)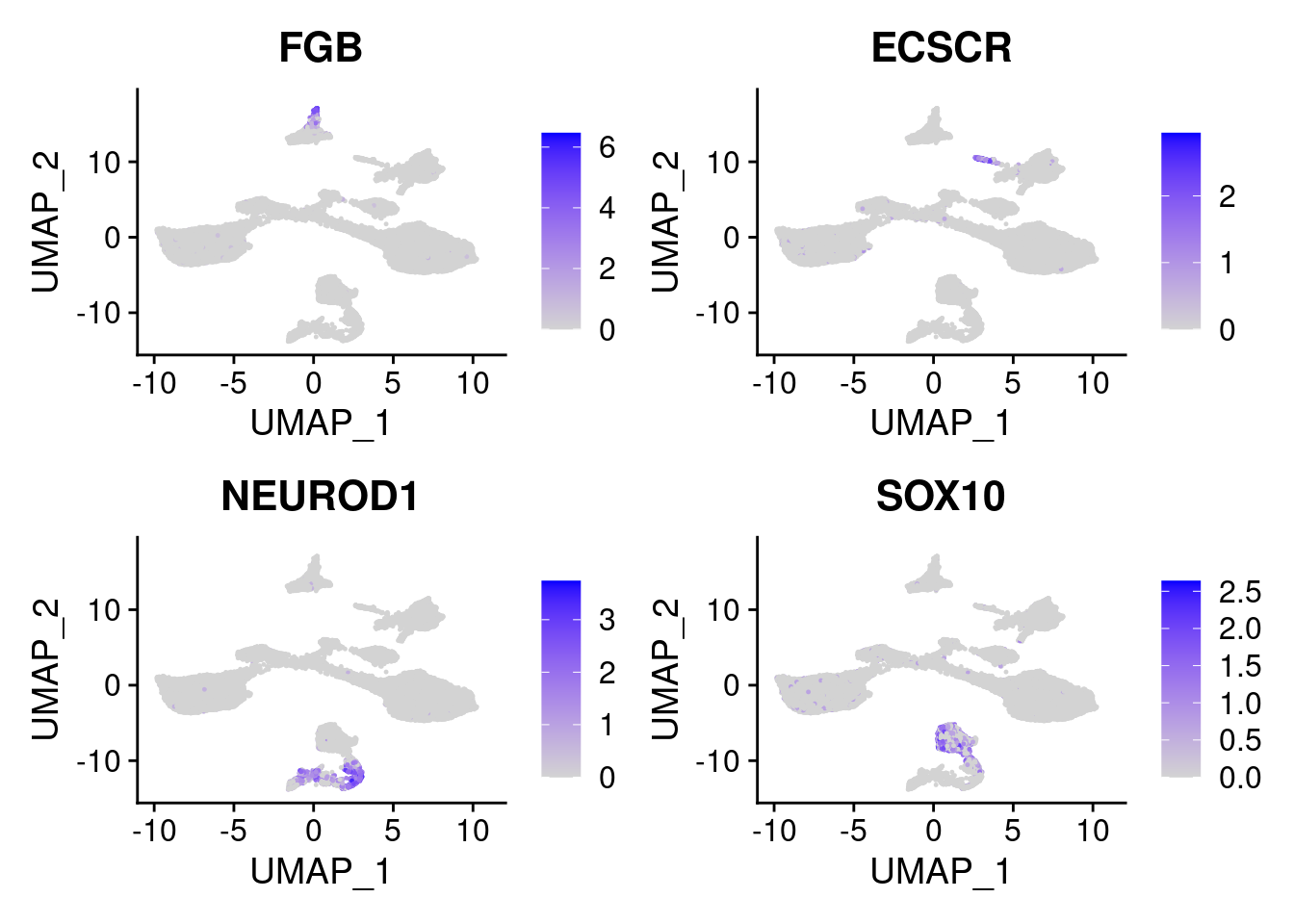
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
sessionInfo()R version 3.6.1 (2019-07-05)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Scientific Linux 7.4 (Nitrogen)
Matrix products: default
BLAS/LAPACK: /software/openblas-0.2.19-el7-x86_64/lib/libopenblas_haswellp-r0.2.19.so
locale:
[1] C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] reshape2_1.4.4 RColorBrewer_1.1-2 here_0.1-11 DataCombine_0.2.21
[5] ggplot2_3.3.2 harmony_1.0 Rcpp_1.0.5 Seurat_3.2.0
[9] workflowr_1.6.2
loaded via a namespace (and not attached):
[1] Rtsne_0.15 colorspace_1.4-1 deldir_0.1-28
[4] ellipsis_0.3.1 ggridges_0.5.2 rprojroot_1.3-2
[7] fs_1.4.2 spatstat.data_1.4-3 farver_2.0.3
[10] leiden_0.3.3 listenv_0.8.0 npsurv_0.4-0
[13] ggrepel_0.8.2 RSpectra_0.16-0 codetools_0.2-16
[16] splines_3.6.1 lsei_1.2-0 knitr_1.29
[19] polyclip_1.10-0 jsonlite_1.7.0 ica_1.0-2
[22] cluster_2.1.0 png_0.1-7 uwot_0.1.8
[25] shiny_1.5.0 sctransform_0.2.1 compiler_3.6.1
[28] httr_1.4.2 backports_1.1.8 Matrix_1.2-18
[31] fastmap_1.0.1 lazyeval_0.2.2 later_1.1.0.1
[34] htmltools_0.5.0 tools_3.6.1 rsvd_1.0.3
[37] igraph_1.2.5 gtable_0.3.0 glue_1.4.1
[40] RANN_2.6.1 dplyr_1.0.0 rappdirs_0.3.1
[43] spatstat_1.64-1 vctrs_0.3.2 gdata_2.18.0
[46] ape_5.3 nlme_3.1-140 lmtest_0.9-37
[49] xfun_0.16 stringr_1.4.0 globals_0.12.5
[52] mime_0.9 miniUI_0.1.1.1 lifecycle_0.2.0
[55] irlba_2.3.3 gtools_3.8.2 goftest_1.2-2
[58] future_1.18.0 MASS_7.3-51.4 zoo_1.8-8
[61] scales_1.1.1 promises_1.1.1 spatstat.utils_1.17-0
[64] parallel_3.6.1 yaml_2.2.1 reticulate_1.16
[67] pbapply_1.4-2 gridExtra_2.3 rpart_4.1-15
[70] stringi_1.4.6 caTools_1.18.0 rlang_0.4.7
[73] pkgconfig_2.0.3 bitops_1.0-6 evaluate_0.14
[76] lattice_0.20-38 ROCR_1.0-7 purrr_0.3.4
[79] tensor_1.5 labeling_0.3 patchwork_1.0.1
[82] htmlwidgets_1.5.1 cowplot_1.0.0 tidyselect_1.1.0
[85] RcppAnnoy_0.0.16 plyr_1.8.6 magrittr_1.5
[88] R6_2.4.1 gplots_3.0.4 generics_0.0.2
[91] withr_2.2.0 pillar_1.4.6 whisker_0.4
[94] mgcv_1.8-28 fitdistrplus_1.0-14 survival_3.2-3
[97] abind_1.4-5 tibble_3.0.3 future.apply_1.6.0
[100] crayon_1.3.4 KernSmooth_2.23-15 plotly_4.9.2.1
[103] rmarkdown_2.3 grid_3.6.1 data.table_1.13.0
[106] git2r_0.26.1 digest_0.6.25 xtable_1.8-4
[109] tidyr_1.1.0 httpuv_1.5.4 munsell_0.5.0
[112] viridisLite_0.3.0