Integrate lanes from all batches of EB pilot
Last updated: 2021-07-05
Checks: 7 0
Knit directory: Embryoid_Body_Pilot_Workflowr/analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it's best to always run the code in an empty environment.
The command set.seed(20200804) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version e9247fb. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: analysis/.Rhistory
Ignored: output/.Rhistory
Untracked files:
Untracked: GSE122380_raw_counts.txt.gz
Untracked: UTF1_plots.Rmd
Untracked: analysis/IntegrateReference_SCTregressCaoPlusScHCL_JustEarlyEcto.Rmd
Untracked: analysis/IntegrateReference_SCTregressCaoPlusScHCL_JustEndo.Rmd
Untracked: analysis/IntegrateReference_SCTregressCaoPlusScHCL_JustMeso.Rmd
Untracked: analysis/IntegrateReference_SCTregressCaoPlusScHCL_JustNeuralCrest.Rmd
Untracked: analysis/IntegrateReference_SCTregressCaoPlusScHCL_JustNeuron.Rmd
Untracked: analysis/IntegrateReference_SCTregressCaoPlusScHCL_JustPluri.Rmd
Untracked: analysis/OLD/
Untracked: analysis/Pseudobulk_Limma_Harmony.BatchIndividual_ClusterRes0.8_minPCT0.2.Rmd
Untracked: analysis/Pseudobulk_Limma_Harmony.BatchIndividual_ClusterRes1_minPCT0.2.Rmd
Untracked: analysis/Pseudobulk_VariancePartition_Harmony.Batchindividual_ClusterRes0.1_byCluster.Rmd
Untracked: analysis/RefInt_ComparingFulltoPartialIntegrationAnnotations.Rmd
Untracked: analysis/ReferenceAnn_DE.Rmd
Untracked: analysis/SingleCell_HierarchicalClustering_NoGeneFilter.Rmd
Untracked: analysis/SingleCell_VariancePartitionByCluster_Harmony.Batchindividual_ClusterRes0.1_minPCT0.2.Rmd
Untracked: analysis/VarPartPlots_res0.1_SCT.Rmd
Untracked: analysis/VarPart_SC_res0.1_SCT.Rmd
Untracked: analysis/child/
Untracked: analysis/k10topics_Explore.Rmd
Untracked: analysis/k6topics_Explore.Rmd
Untracked: build_refint_scale.R
Untracked: build_refint_sct.R
Untracked: build_stuff.R
Untracked: build_varpart_sc.R
Untracked: code/.ipynb_checkpoints/
Untracked: code/CellRangerPreprocess.Rmd
Untracked: code/ConvertToDGE.Rmd
Untracked: code/ConvertToDGE_PseudoBulk.Rmd
Untracked: code/ConvertToDGE_SingleCellRes_minPCT0.2.Rmd
Untracked: code/EB.getHumanMetadata.Rmd
Untracked: code/GEO_processed_data.Rmd
Untracked: code/PowerAnalysis_NoiseRatio.ipynb
Untracked: code/Untitled.ipynb
Untracked: code/Untitled1.ipynb
Untracked: code/compile_fits.Rmd
Untracked: code/fit_all_models.sh
Untracked: code/fit_poisson_nmf.R
Untracked: code/fit_poisson_nmf.sbatch
Untracked: code/functions_for_fit_comparison.Rmd
Untracked: code/get_genelist_byPCTthresh.Rmd
Untracked: code/prefit_poisson_nmf.R
Untracked: code/prefit_poisson_nmf.sbatch
Untracked: code/prepare_data_for_fastTopics.Rmd
Untracked: data/HCL_Fig1_adata.h5ad
Untracked: data/HCL_Fig1_adata.h5seurat
Untracked: data/dge/
Untracked: data/dge_raw_data.tar.gz
Untracked: data/ref.expr.rda
Untracked: figure/
Untracked: output/CR_sampleQCrds/
Untracked: output/CaoEtAl.Obj.CellsOfAllClusters.ProteinCodingGenes.rds
Untracked: output/CaoEtAl.Obj.rds
Untracked: output/ClusterInfo_res0.1.csv
Untracked: output/DGELists/
Untracked: output/DownSampleVarPart.rds
Untracked: output/Frequency.MostCommonAnnotation.FiveNearestRefCells.csv
Untracked: output/GEOsubmissionProcessedFiles/
Untracked: output/GeneLists_by_minPCT/
Untracked: output/MostCommonAnnotation.FiveNearestRefCells.csv
Untracked: output/NearestReferenceCell.Cao.hESC.EuclideanDistanceinHarmonySpace.csv
Untracked: output/NearestReferenceCell.Cao.hESC.FrequencyofEachAnnotation.csv
Untracked: output/NearestReferenceCell.SCTregressRNAassay.Cao.hESC.EuclideanDistanceinHarmonySpace.csv
Untracked: output/NearestReferenceCell.SCTregressRNAassay.Cao.hESC.FrequencyofEachAnnotation.csv
Untracked: output/Pseudobulk_Limma_res0.1_OnevAllTopTables.csv
Untracked: output/Pseudobulk_Limma_res0.1_OnevAll_top10Upregby_adjP.csv
Untracked: output/Pseudobulk_Limma_res0.1_OnevAll_top10Upregby_logFC.csv
Untracked: output/Pseudobulk_Limma_res0.5_OnevAllTopTables.csv
Untracked: output/Pseudobulk_Limma_res0.8_OnevAllTopTables.csv
Untracked: output/Pseudobulk_Limma_res1_OnevAllTopTables.csv
Untracked: output/Pseudobulk_VarPart.ByCluster.Res0.1.rds
Untracked: output/ResidualVariances_fromDownSampAnalysis.csv
Untracked: output/SingleCell_VariancePartition_RNA_Res0.1_minPCT0.2.rds
Untracked: output/SingleCell_VariancePartition_Res0.1_minPCT0.2.rds
Untracked: output/SingleCell_VariancePartition_SCT_Res0.1_minPCT0.2.rds
Untracked: output/TopicModelling_k10_top10drivergenes.byBeta.csv
Untracked: output/TopicModelling_k6_top10drivergenes.byBeta.csv
Untracked: output/TopicModelling_k6_top15drivergenes.byZ.csv
Untracked: output/TranferredAnnotations_ReferenceInt_JustEarlyEcto.csv
Untracked: output/TranferredAnnotations_ReferenceInt_JustEndoderm.csv
Untracked: output/TranferredAnnotations_ReferenceInt_JustMeso.csv
Untracked: output/TranferredAnnotations_ReferenceInt_JustNeuralCrest.csv
Untracked: output/TranferredAnnotations_ReferenceInt_JustNeuron.csv
Untracked: output/TranferredAnnotations_ReferenceInt_JustPluripotent.csv
Untracked: output/VarPart.ByCluster.Res0.1.rds
Untracked: output/azimuth/
Untracked: output/downsamp_10800cells_10subreps_medianexplainedbyresiduals_varpart_PsB.rds
Untracked: output/downsamp_16200cells_10subreps_medianexplainedbyresiduals_varpart_PsB.rds
Untracked: output/downsamp_21600cells_10subreps_medianexplainedbyresiduals_varpart_PsB.rds
Untracked: output/downsamp_2700cells_10subreps_medianexplainedbyresiduals_varpart_PsB.rds
Untracked: output/downsamp_2700cells_10subreps_medianexplainedbyresiduals_varpart_scres.rds
Untracked: output/downsamp_5400cells_10subreps_medianexplainedbyresiduals_varpart_PsB.rds
Untracked: output/downsamp_7200cells_10subreps_medianexplainedbyresiduals_varpart_PsB.rds
Untracked: output/fasttopics/
Untracked: output/figs/
Untracked: output/merge.Cao.SCTwRegressOrigIdent.rds
Untracked: output/merge.all.SCTwRegressOrigIdent.Harmony.rds
Untracked: output/merged.SCT.counts.matrix.rds
Untracked: output/merged.raw.counts.matrix.rds
Untracked: output/mergedObjects/
Untracked: output/pdfs/
Untracked: output/sampleQCrds/
Untracked: output/splitgpm_gsea_results/
Untracked: slurm-12005914.out
Untracked: slurm-12005923.out
Unstaged changes:
Deleted: analysis/IntegrateAnalysis.afterFilter.HarmonyBatch.Rmd
Deleted: analysis/IntegrateAnalysis.afterFilter.HarmonyBatchSampleIDindividual.Rmd
Deleted: analysis/IntegrateAnalysis.afterFilter.NOHARMONYjustmerge.Rmd
Deleted: analysis/IntegrateAnalysis.afterFilter.SCTregressBatchIndividual.Rmd
Deleted: analysis/IntegrateAnalysis.afterFilter.SCTregressBatchIndividualHarmonyBatchindividual.Rmd
Modified: analysis/Pseudobulk_HierarchicalClustering_Harmony.Batchindividual_ClusterRes0.1_minPCT0.2.Rmd
Modified: analysis/Pseudobulk_HierarchicalClustering_Harmony.Batchindividual_ClusterRes0.5_minPCT0.2.Rmd
Modified: analysis/Pseudobulk_HierarchicalClustering_Harmony.Batchindividual_ClusterRes0.8_minPCT0.2.Rmd
Modified: analysis/Pseudobulk_HierarchicalClustering_Harmony.Batchindividual_ClusterRes1_minPCT0.2.Rmd
Modified: analysis/Pseudobulk_Limma_Harmony.BatchIndividual_ClusterRes0.1_minPCT0.2.Rmd
Modified: analysis/Pseudobulk_Limma_Harmony.BatchIndividual_ClusterRes0.5_minPCT0.2.Rmd
Modified: analysis/Pseudobulk_VariancePartition_Harmony.Batchindividual_ClusterRes0.1_minPCT0.2.Rmd
Modified: analysis/Pseudobulk_VariancePartition_Harmony.Batchindividual_ClusterRes0.5_minPCT0.2.Rmd
Modified: analysis/Pseudobulk_VariancePartition_Harmony.Batchindividual_ClusterRes0.8_minPCT0.2.Rmd
Modified: analysis/Pseudobulk_VariancePartition_Harmony.Batchindividual_ClusterRes1_minPCT0.2.Rmd
Deleted: analysis/RunscHCL_HarmonyBatchInd.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/IntegrateAnalysis.afterFilter.HarmonyBatchindividual.Rmd) and HTML (docs/IntegrateAnalysis.afterFilter.HarmonyBatchindividual.html) files. If you've configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | e9247fb | KLRhodes | 2021-07-05 | wflow_publish("analysis/IntegrateAnalysis.afterFilter.HarmonyBatchindividual.Rmd") |
| html | edaa6a3 | KLRhodes | 2020-08-11 | Build site. |
| Rmd | f50ebd3 | KLRhodes | 2020-08-10 | wflow_publish("analysis/Integrate*") |
| html | 421a225 | KLRhodes | 2020-08-10 | Build site. |
| Rmd | bc8ec6f | KLRhodes | 2020-08-10 | cleaning various versions of merging/intCurrent working directory |
library(Seurat)
library(harmony)
library(ggplot2)
library(DataCombine)
library(here)
library(RColorBrewer)
options(future.globals.maxSize= 15000*1024^2) #allow global exceeding 4GbRead in the files, add metadata, and create an object list
filelist<-list.files(here::here('output/sampleQCrds/'), full.names = T)
objectlist<- list()
for (i in 1:length(filelist)){
rds<- readRDS(filelist[i])
objectlist[i]<- rds
}create a merged seurat object
ids<-substr(basename(filelist),1,12)
merged<- merge(objectlist[[1]], c(objectlist[[2]], objectlist[[3]],objectlist[[4]],objectlist[[5]],objectlist[[6]],objectlist[[7]],objectlist[[8]],objectlist[[9]],objectlist[[10]],objectlist[[11]],objectlist[[12]],objectlist[[13]],objectlist[[14]],objectlist[[15]],objectlist[[16]]),add.cell.ids=ids, merge.data=T)#need to fix the individual names because they are slightly different from batch1
replacements<- data.frame(from= c("SNG-NA18511.variant2", "SNG-NA18858.variant2", "SNG-NA19160.variant2"), to=c("SNG-NA18511", "SNG-NA18858", "SNG-NA19160"))
merged@meta.data<-FindReplace(merged@meta.data, "individual", replacements, from = "from", to= "to", exact=T, vector=F )Only exact matches will be replaced.#run PCA on full dataset pre-alignment
all.genes= rownames(merged)
merged<-FindVariableFeatures(merged,selection.method="vst", nfeatures = 5000)
#have previously used all genes (nfeatures=25000) and clustering by individual rather than batch (based on proportion of cells per cluster) was still observed downstream. Now using 5000 because it is the upper bound of what has been recommended in the literature.
merged<- ScaleData(merged, features = all.genes)Centering and scaling data matrixmerged<-RunPCA(merged, npcs = 100, verbose=F)DimPlot(merged, reduction = "pca", group.by = "Batch")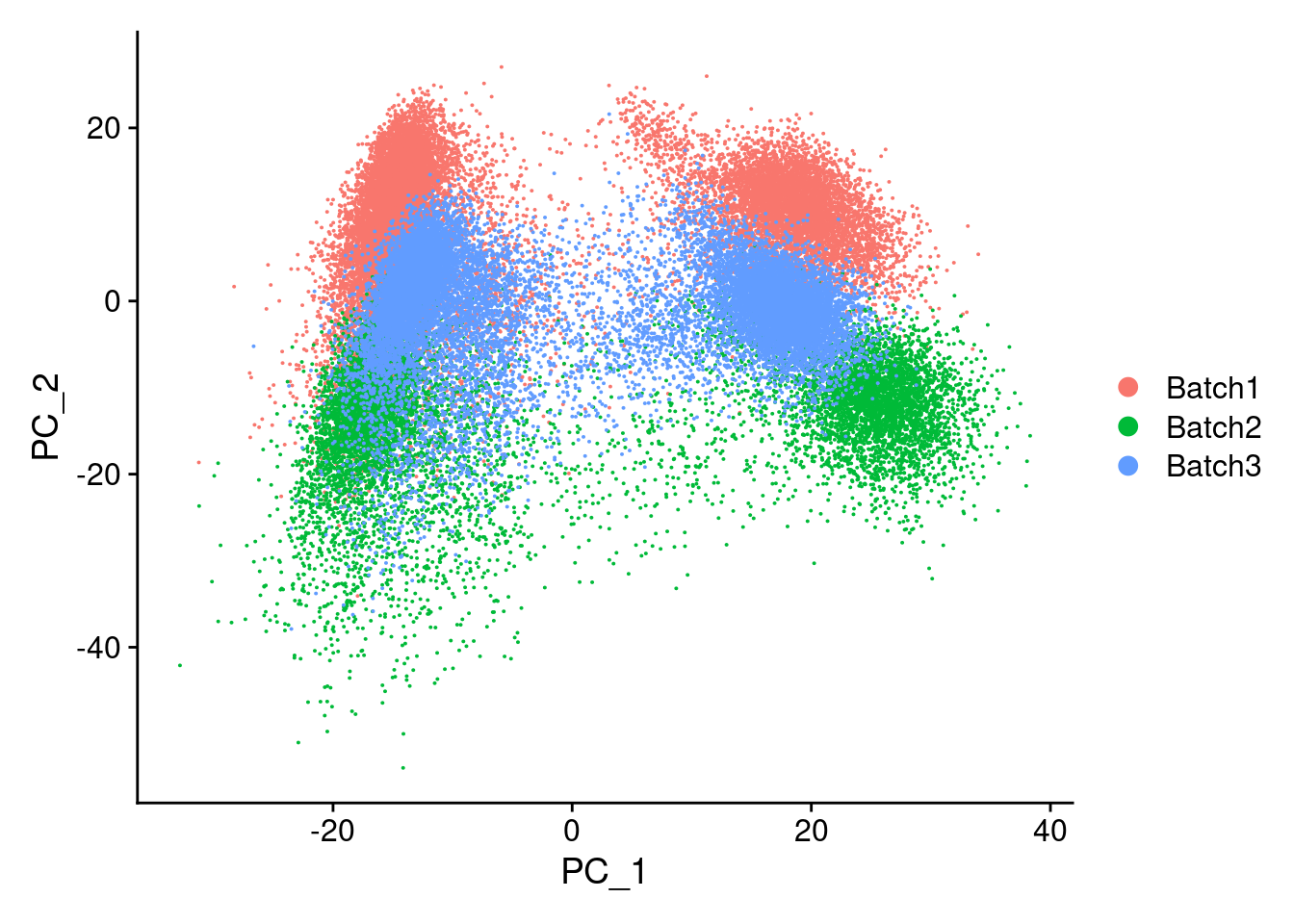
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
Now, running harmony to integrate. Here, using Batch, SampleID(10x Lane), and individual to integrate. Since Batch and Lane are confounded, this may over correct for Batch.
merged<- RunHarmony(merged, c("Batch", "individual"), plot_convergence = T, assay.use = "SCT")Harmony 1/10Harmony 2/10Harmony 3/10Harmony converged after 3 iterationsWarning: Invalid name supplied, making object name syntactically valid. New
object name is Seurat..ProjectDim.SCT.harmony; see ?make.names for more details
on syntax validity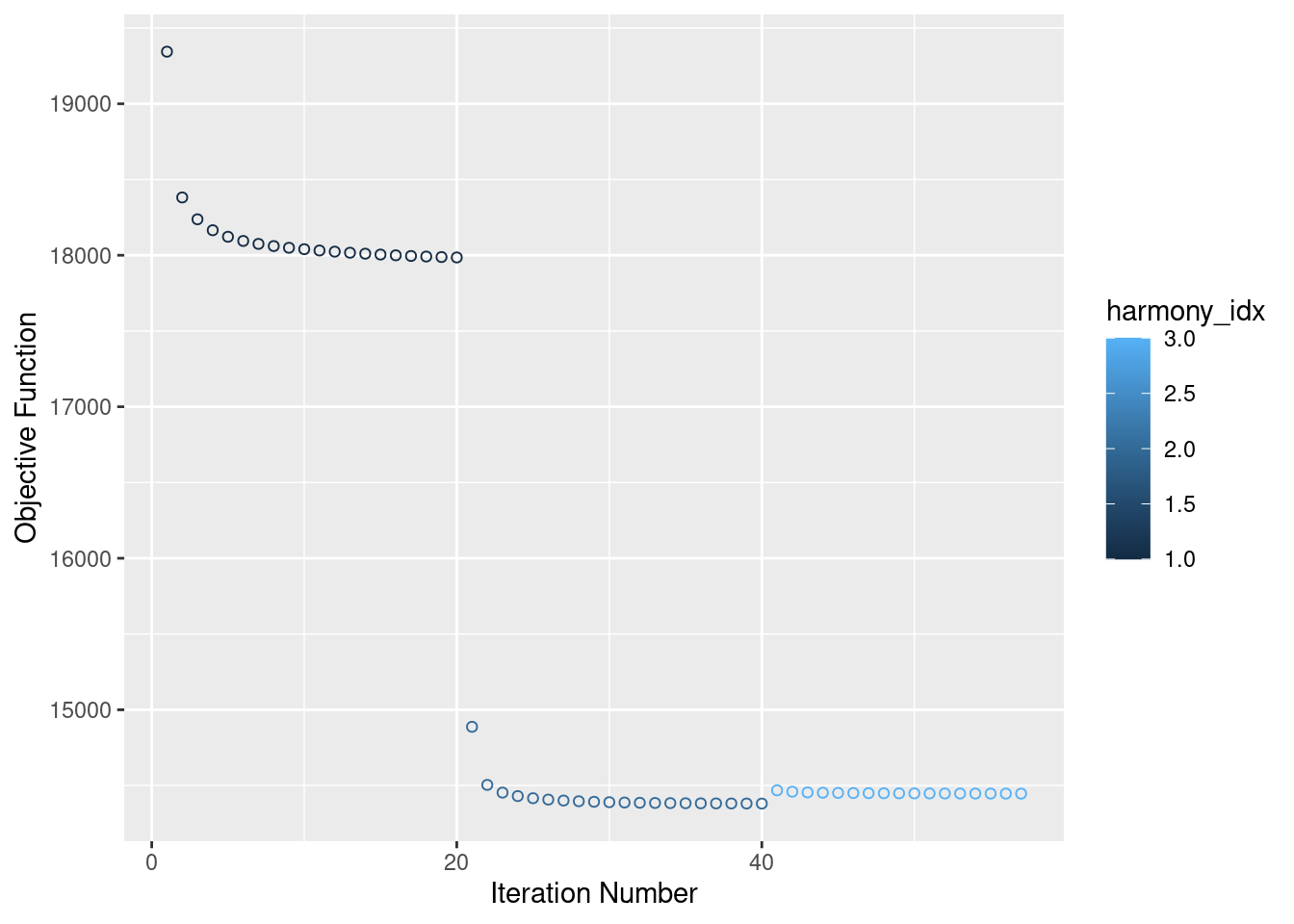
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
Visualize Harmony embeddings
DimPlot(merged, reduction="harmony", group.by= c("individual", "Batch"), combine=F)[[1]]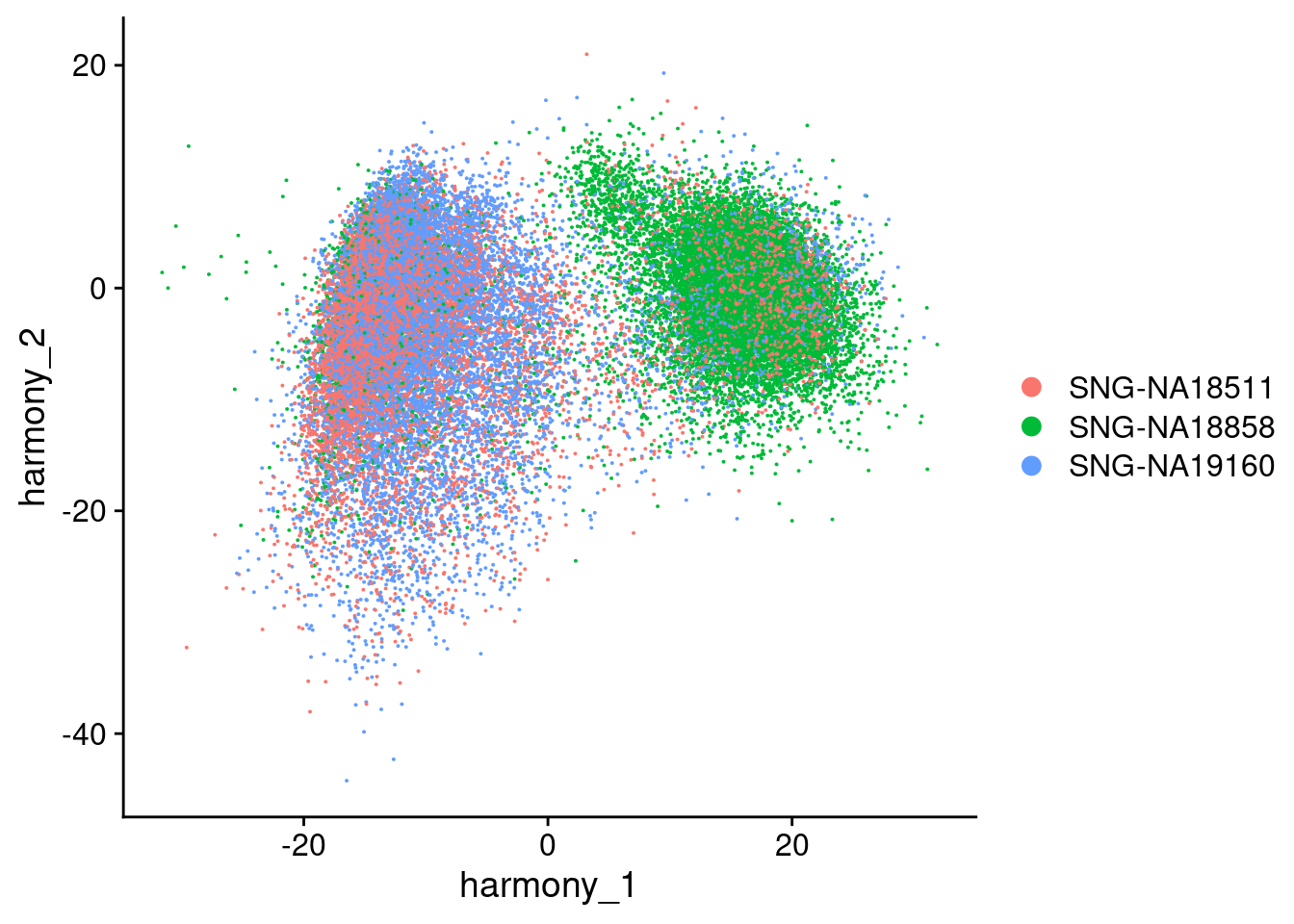
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
[[2]]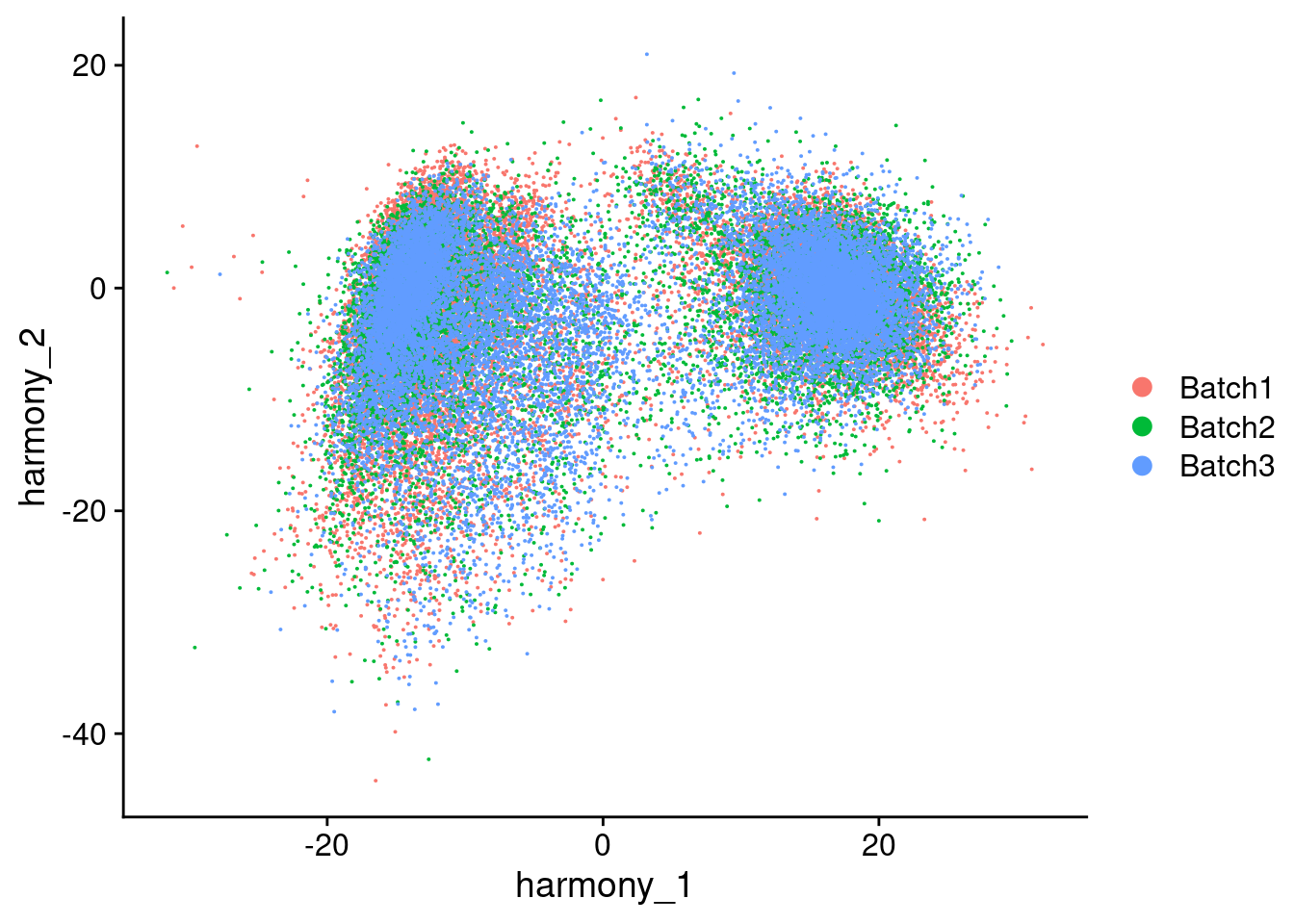
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
Now Running UMAP and identifying clusters, etc
merged<- RunUMAP(merged, reduction = "harmony", dims = 1:100, verbose = F)Warning: The default method for RunUMAP has changed from calling Python UMAP via reticulate to the R-native UWOT using the cosine metric
To use Python UMAP via reticulate, set umap.method to 'umap-learn' and metric to 'correlation'
This message will be shown once per sessionmerged<- FindNeighbors(merged, reduction="harmony", dims = 1:100, verbose = F)
merged<- FindClusters(merged, resolution=1, verbose = F)
merged<- FindClusters(merged, resolution=0.8, verbose = F)
merged<- FindClusters(merged, resolution=0.5, verbose = F)
merged<- FindClusters(merged, resolution=0.1, verbose = F)SAVING merged/aligned/reclustered object
path<- here::here("output/mergedObjects/")
saveRDS(merged, file=paste0(path,'Harmony.Batchindividual.rds'))#reassign idents
Idents(merged)<- 'SCT_snn_res.1'VizDimLoadings(merged, dims = 1:2, reduction = "harmony")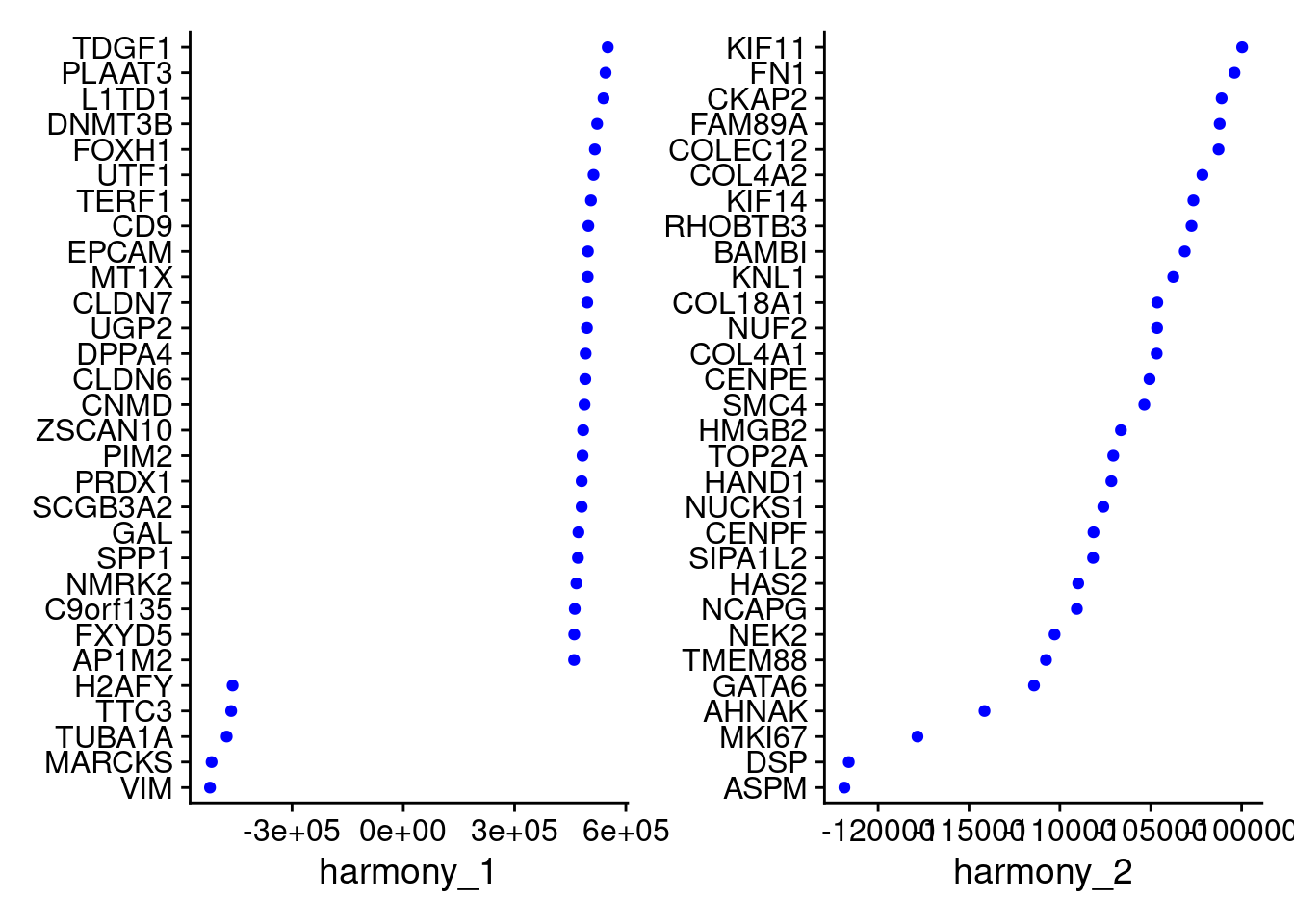
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
VizDimLoadings(merged, dims = 3:4, reduction = "harmony")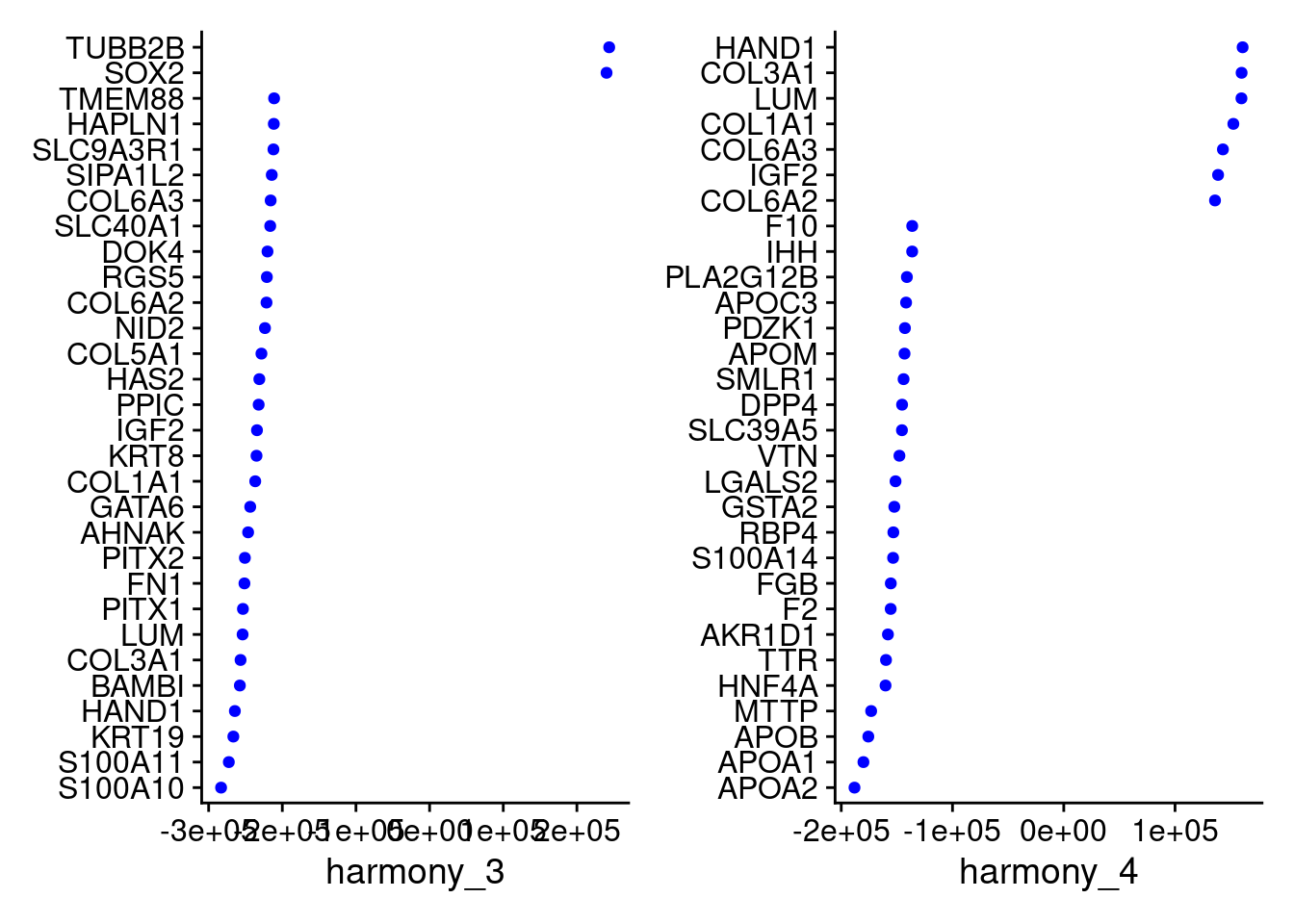
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
VizDimLoadings(merged, dims = 5:6, reduction = "harmony")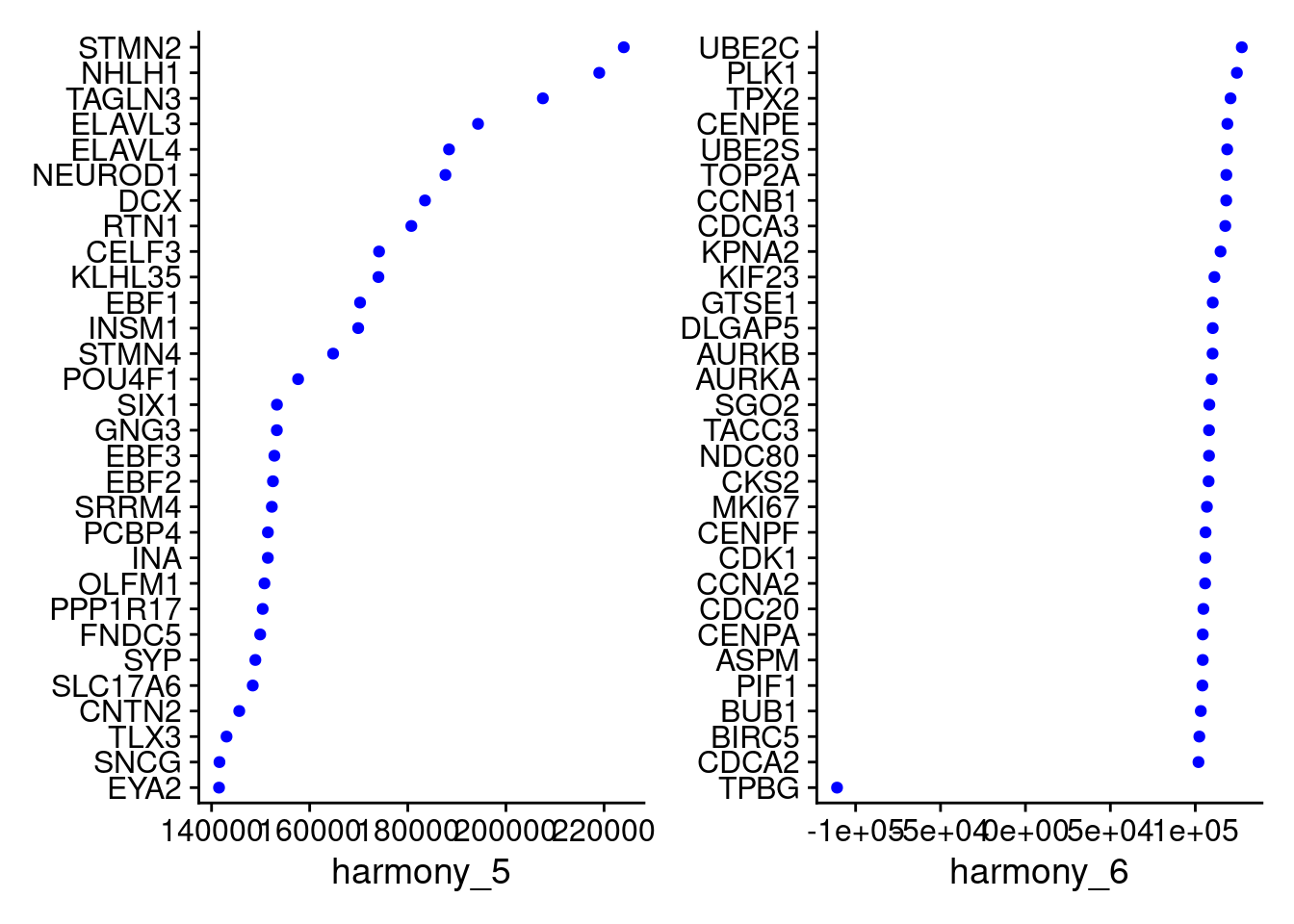
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
xlim <- c(min(merged@reductions$harmony@cell.embeddings[,'harmony_1']),
max(merged@reductions$harmony@cell.embeddings[,'harmony_1']))
ylim <- c(min(merged@reductions$harmony@cell.embeddings[,'harmony_2']),
max(merged@reductions$harmony@cell.embeddings[,'harmony_2']))
individuals <- table(merged$individual)
individuals <- individuals[individuals>50]
individuals <- names(individuals)
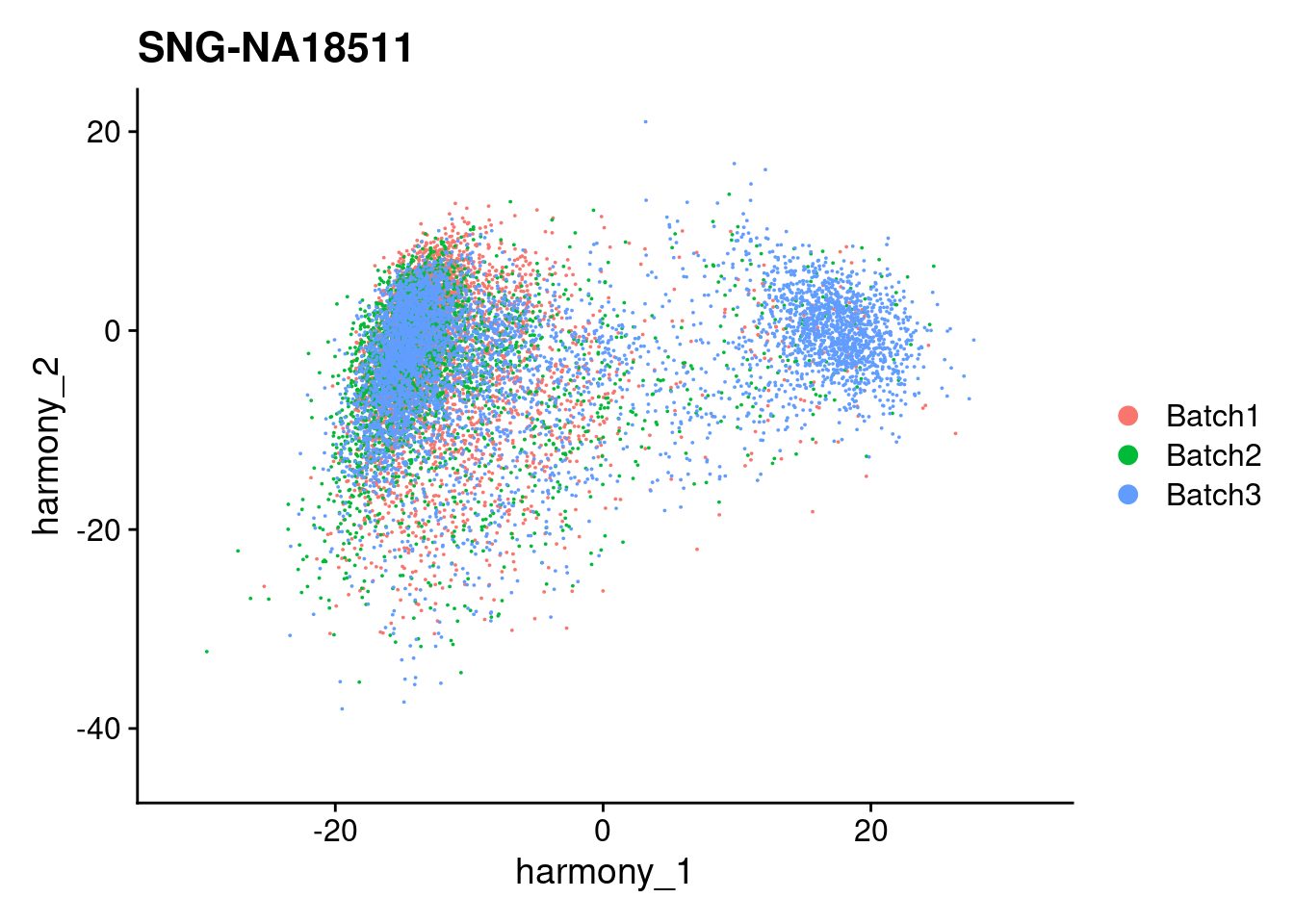
for (i in individuals)
{
print(DimPlot(merged, reduction = "harmony", group.by = c("Batch"), pt.size = 0.01,
cells = WhichCells(merged, expression = individual == i)) +
xlim(xlim) + ylim(ylim) + ggtitle(i))
}
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
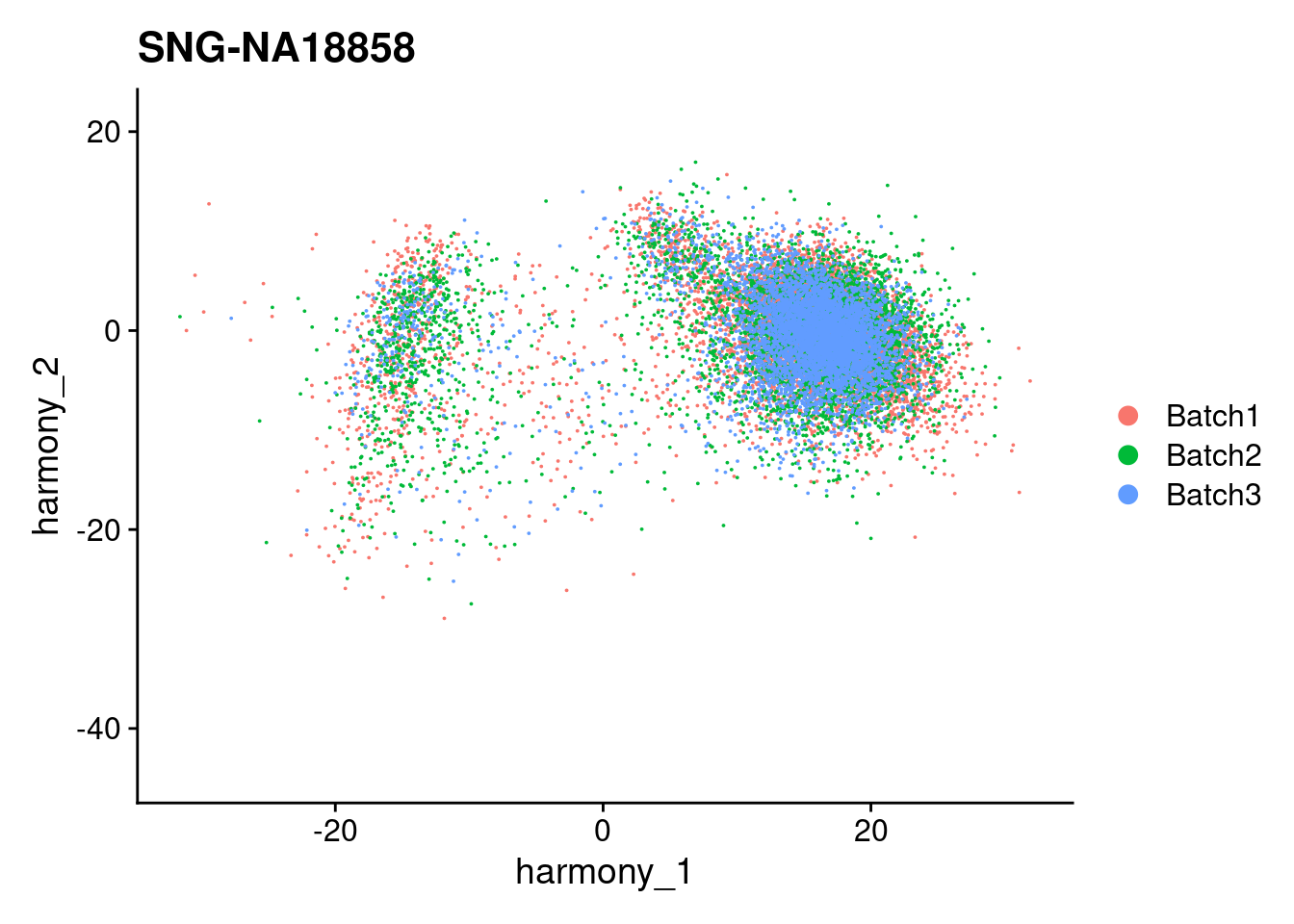
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
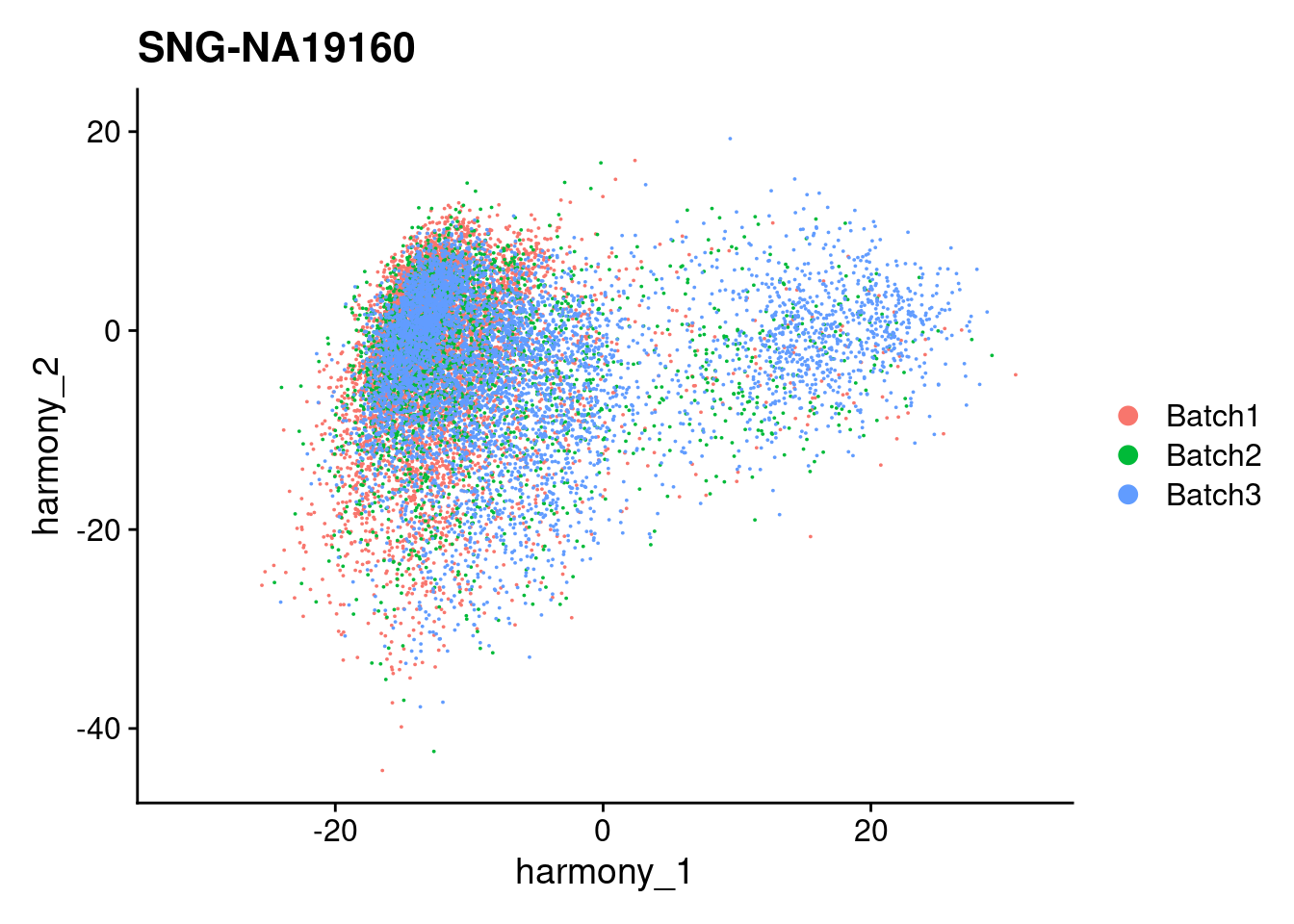
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
DimPlot(merged, reduction = "umap")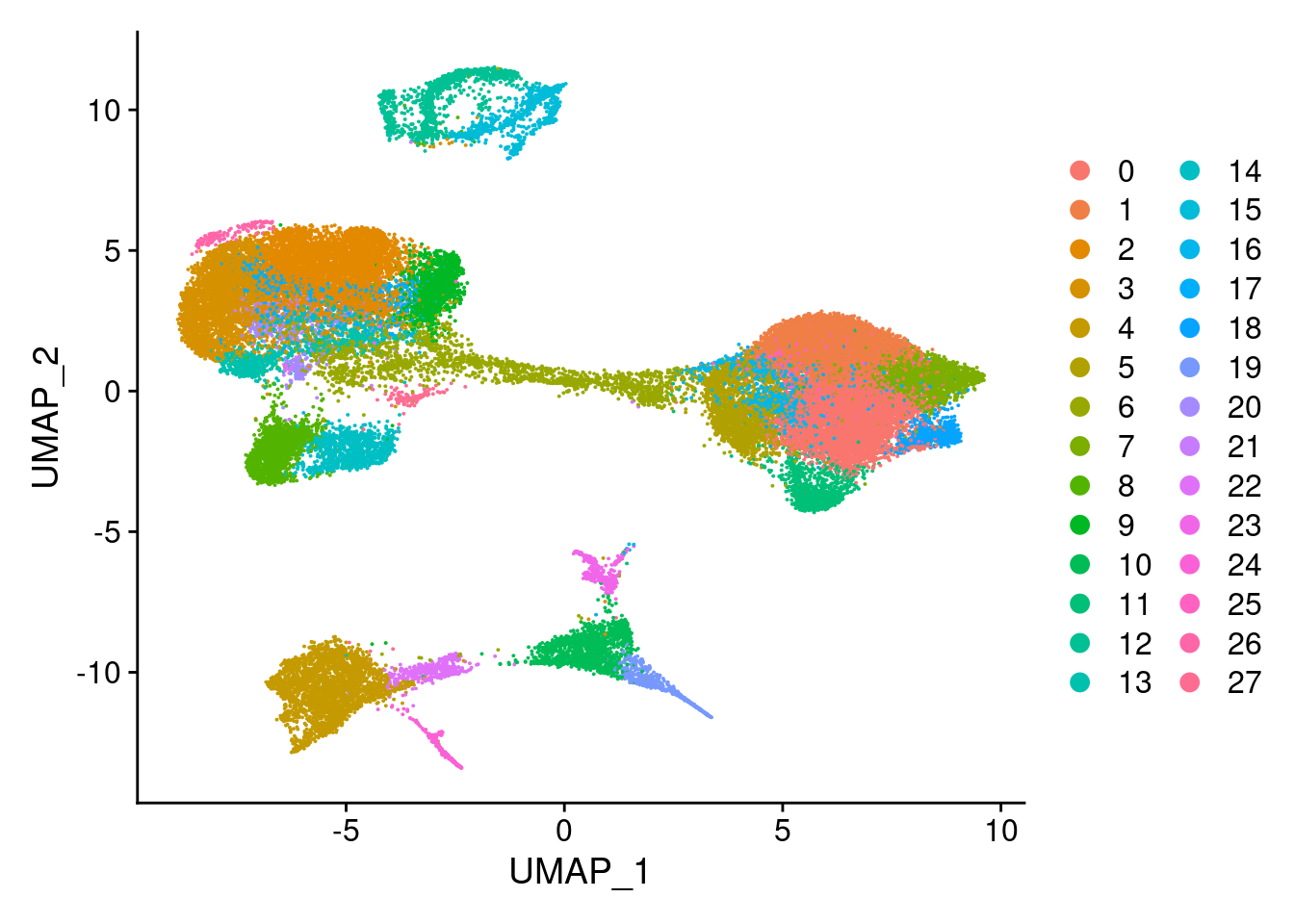
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
DimPlot(merged, reduction = "umap", group.by = "Batch")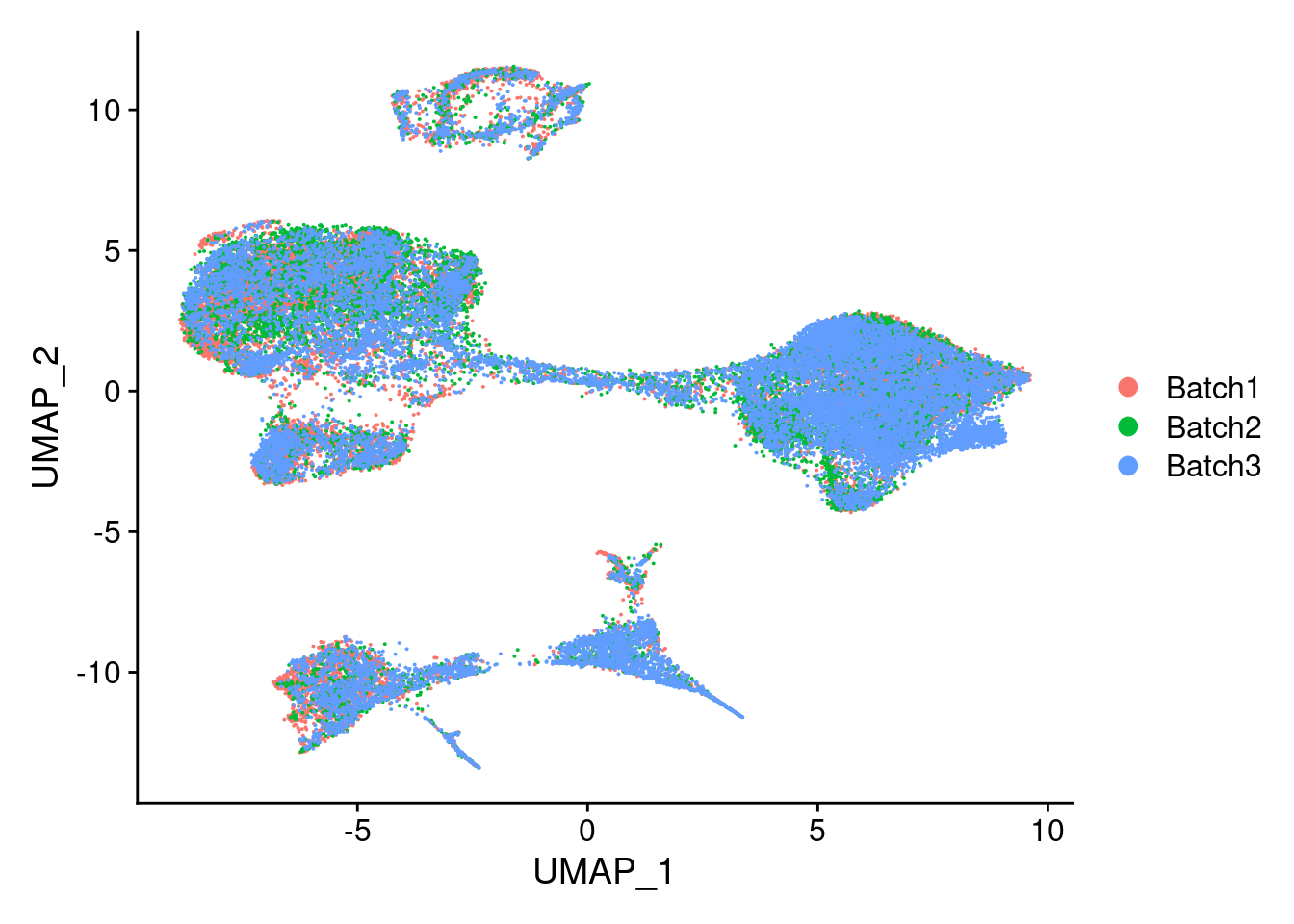
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
DimPlot(merged, reduction = "umap", group.by = "individual")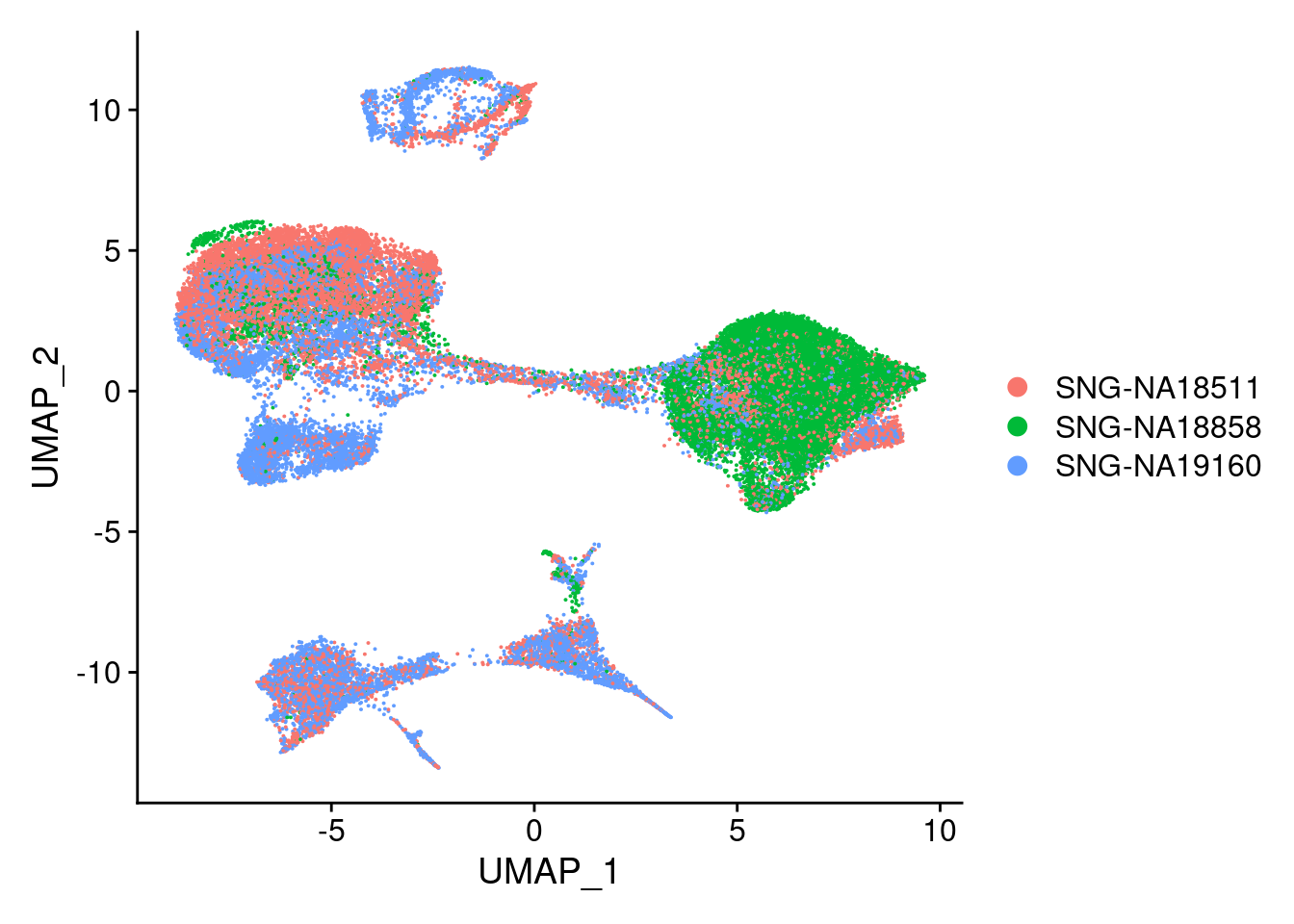
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
xlim <- c(min(merged@reductions$umap@cell.embeddings[,'UMAP_1']),
max(merged@reductions$umap@cell.embeddings[,'UMAP_1']))
ylim <- c(min(merged@reductions$umap@cell.embeddings[,'UMAP_2']),
max(merged@reductions$umap@cell.embeddings[,'UMAP_2']))
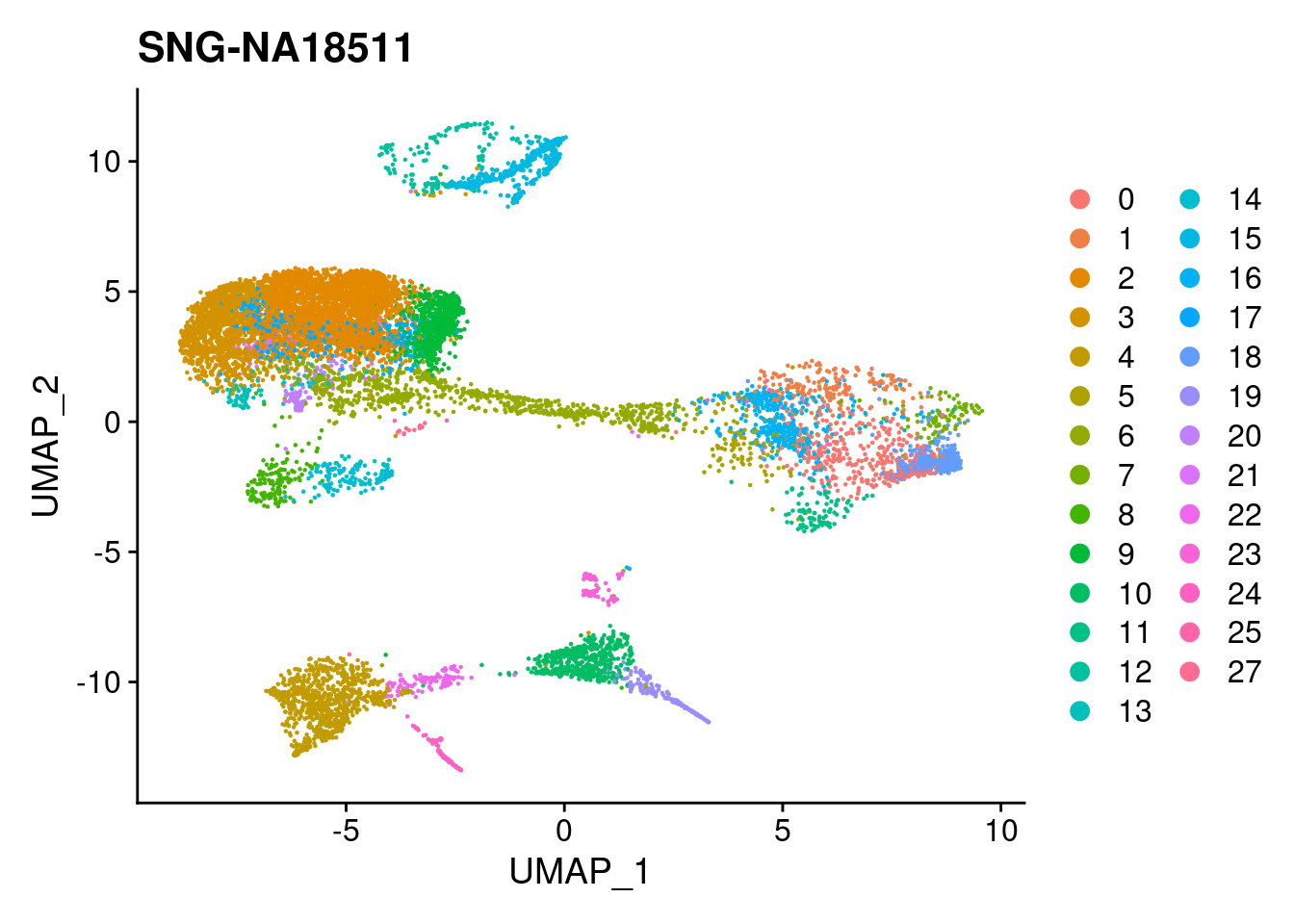
for (i in individuals)
{
print(DimPlot(merged, reduction = "umap",
cells = WhichCells(merged, expression = individual == i)) +
xlim(xlim) + ylim(ylim) + ggtitle(i))
}
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
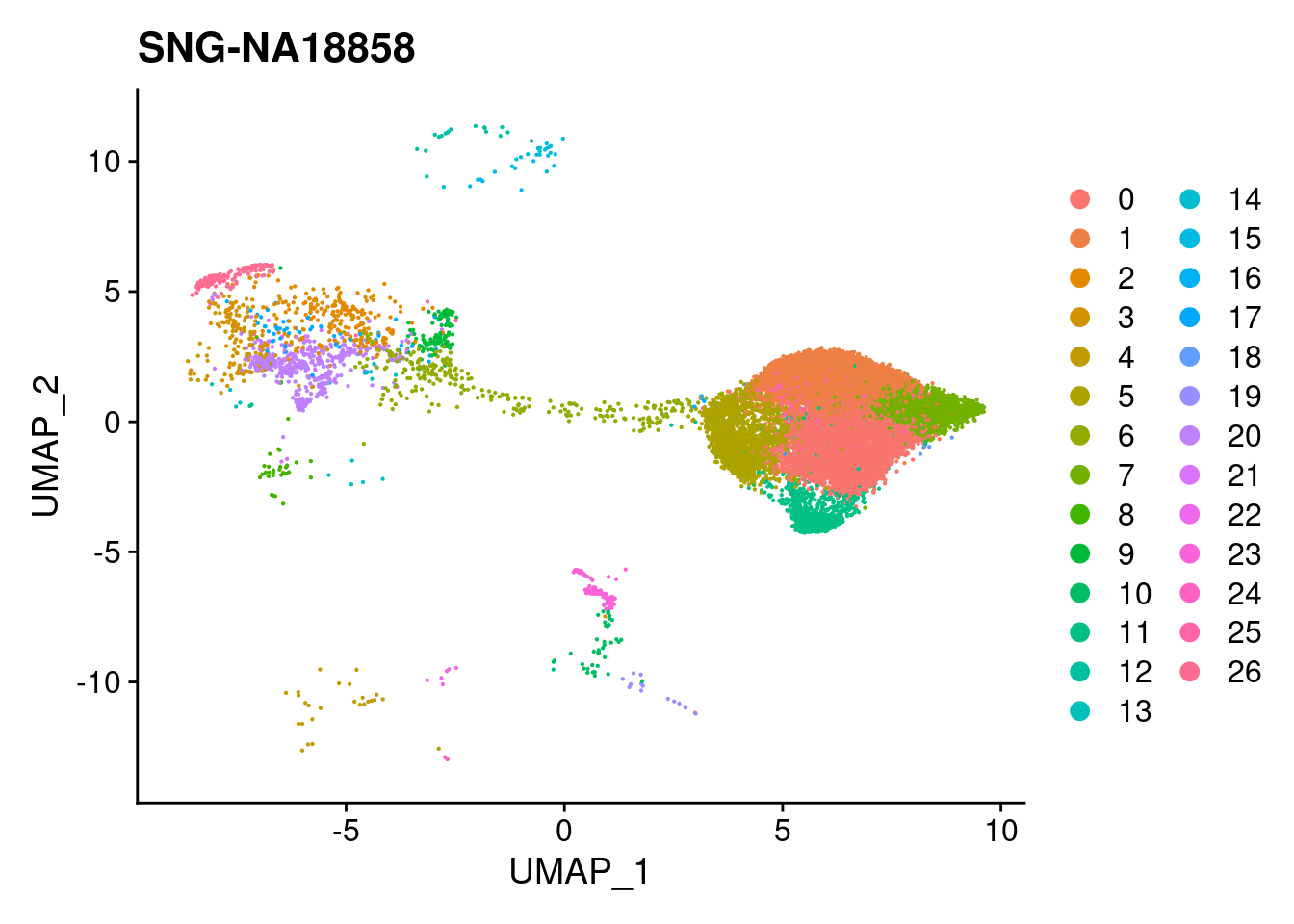
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
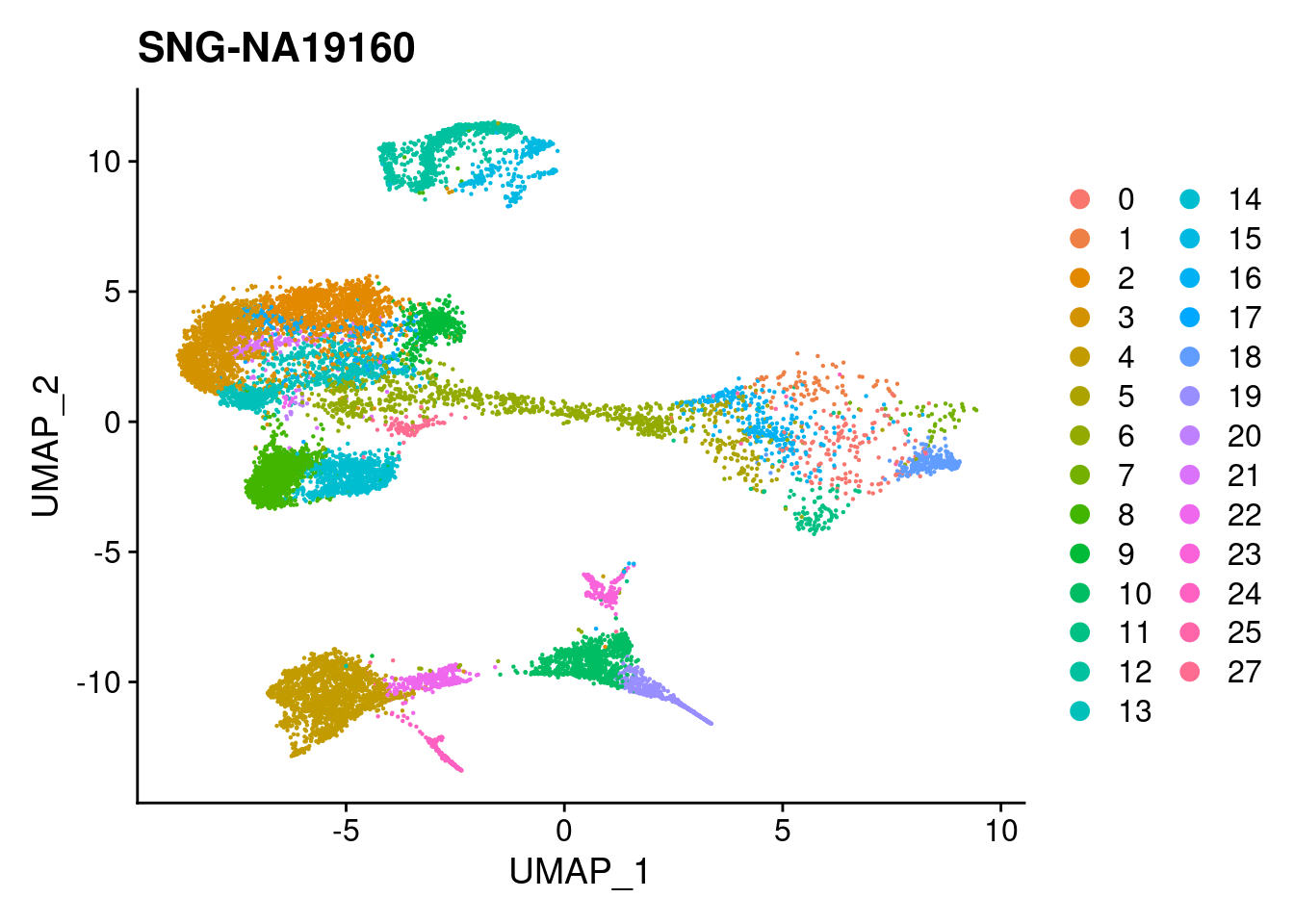
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
plots2<- DimPlot(merged, group.by = "individual", split.by = "Batch")
plots2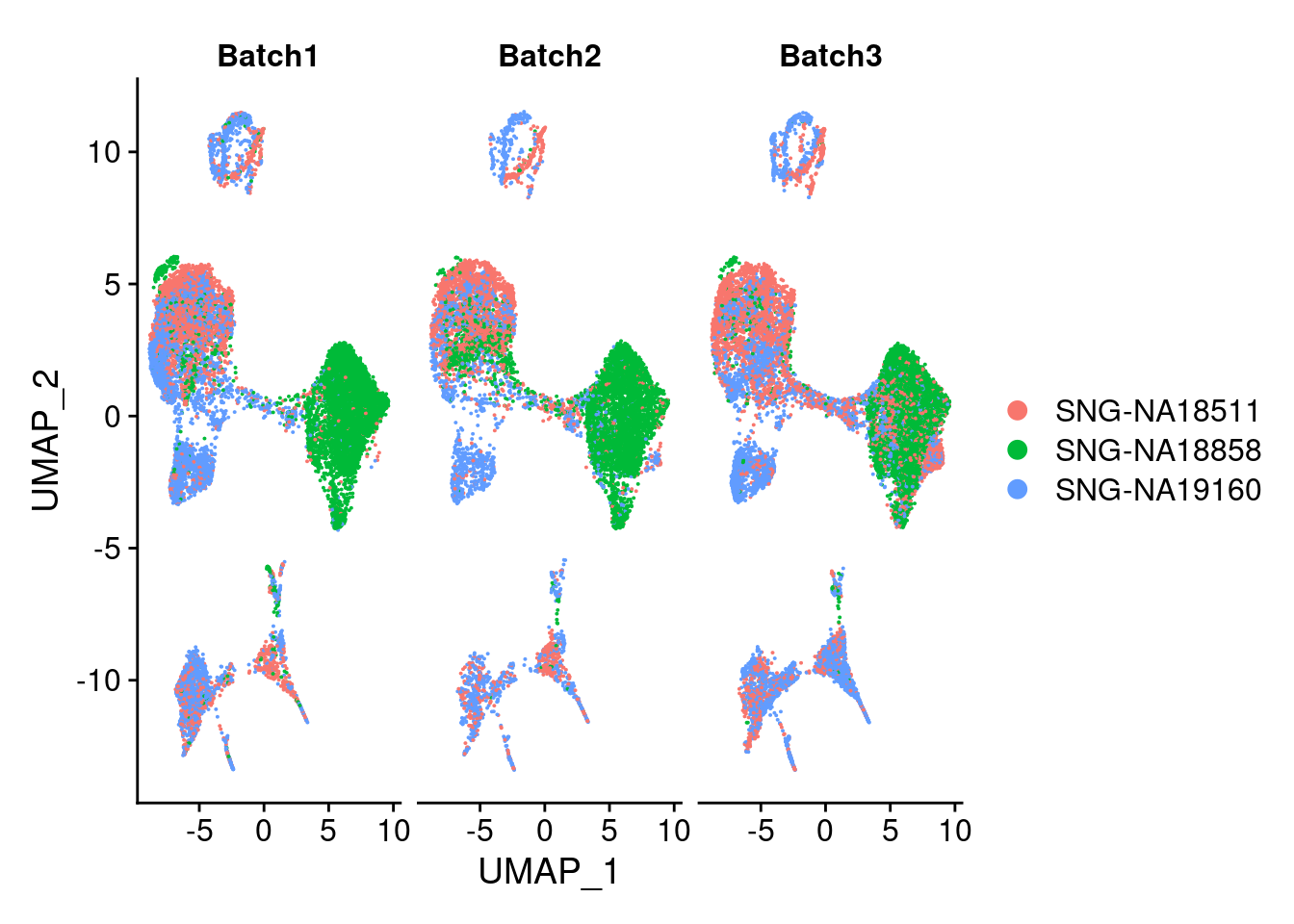
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
DimPlot(merged, group.by = "Batch", split.by = c("individual"))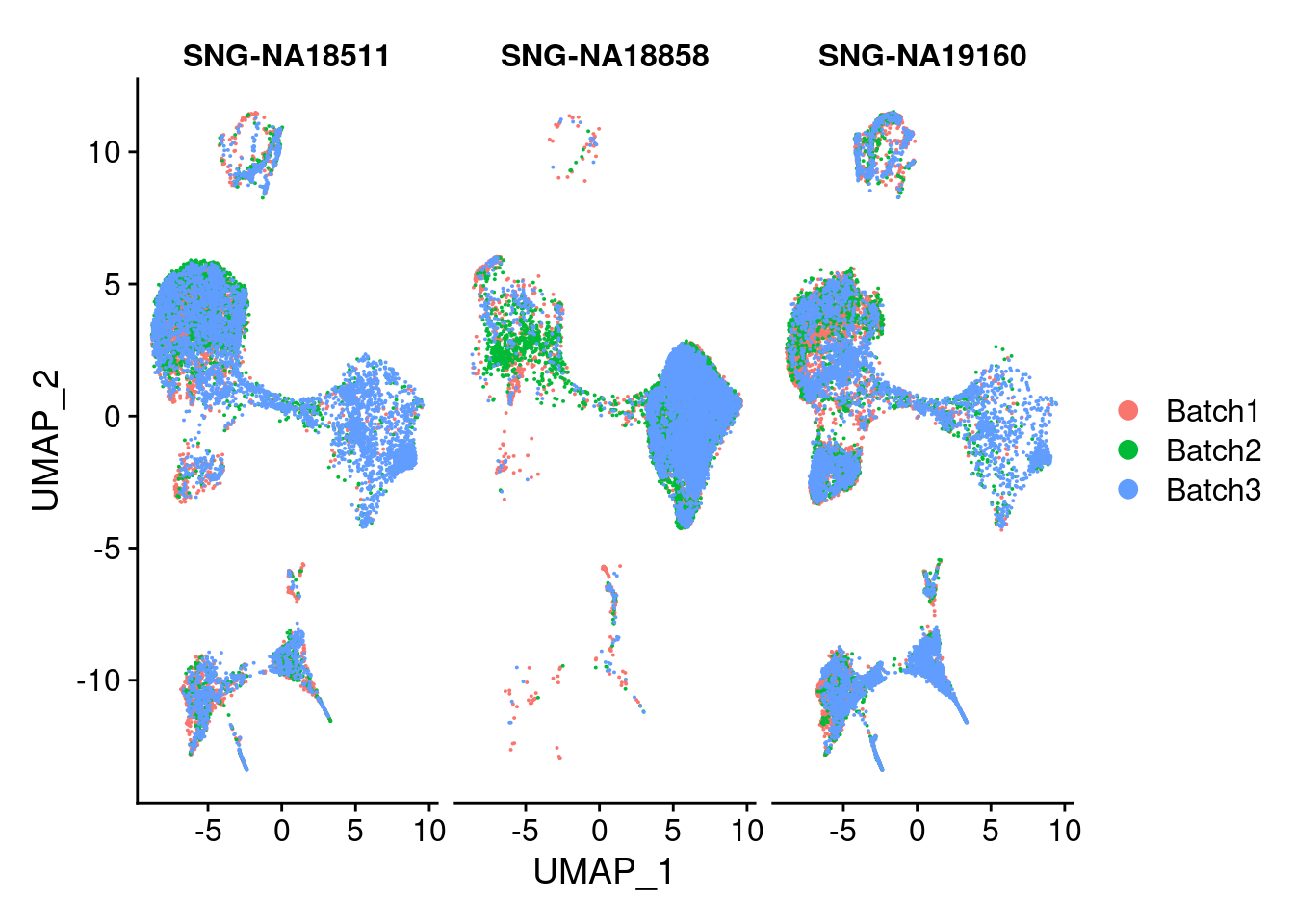
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
DimPlot(merged, group.by = "SCT_snn_res.1", split.by = c("Batch"), label=T)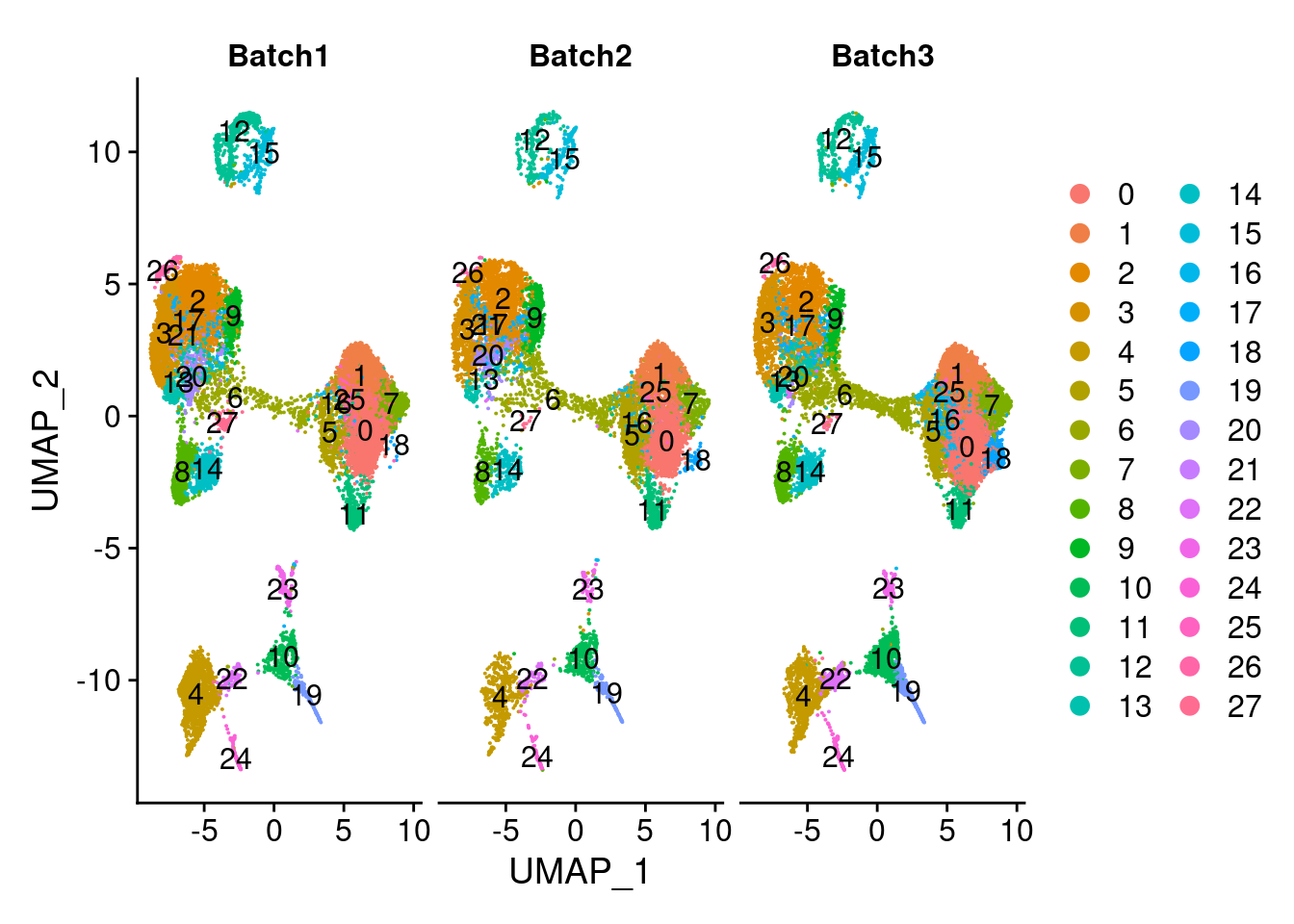
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
DimPlot(merged, reduction = "harmony", group.by = "SCT_snn_res.1", split.by = "Batch", combine = F)[[1]]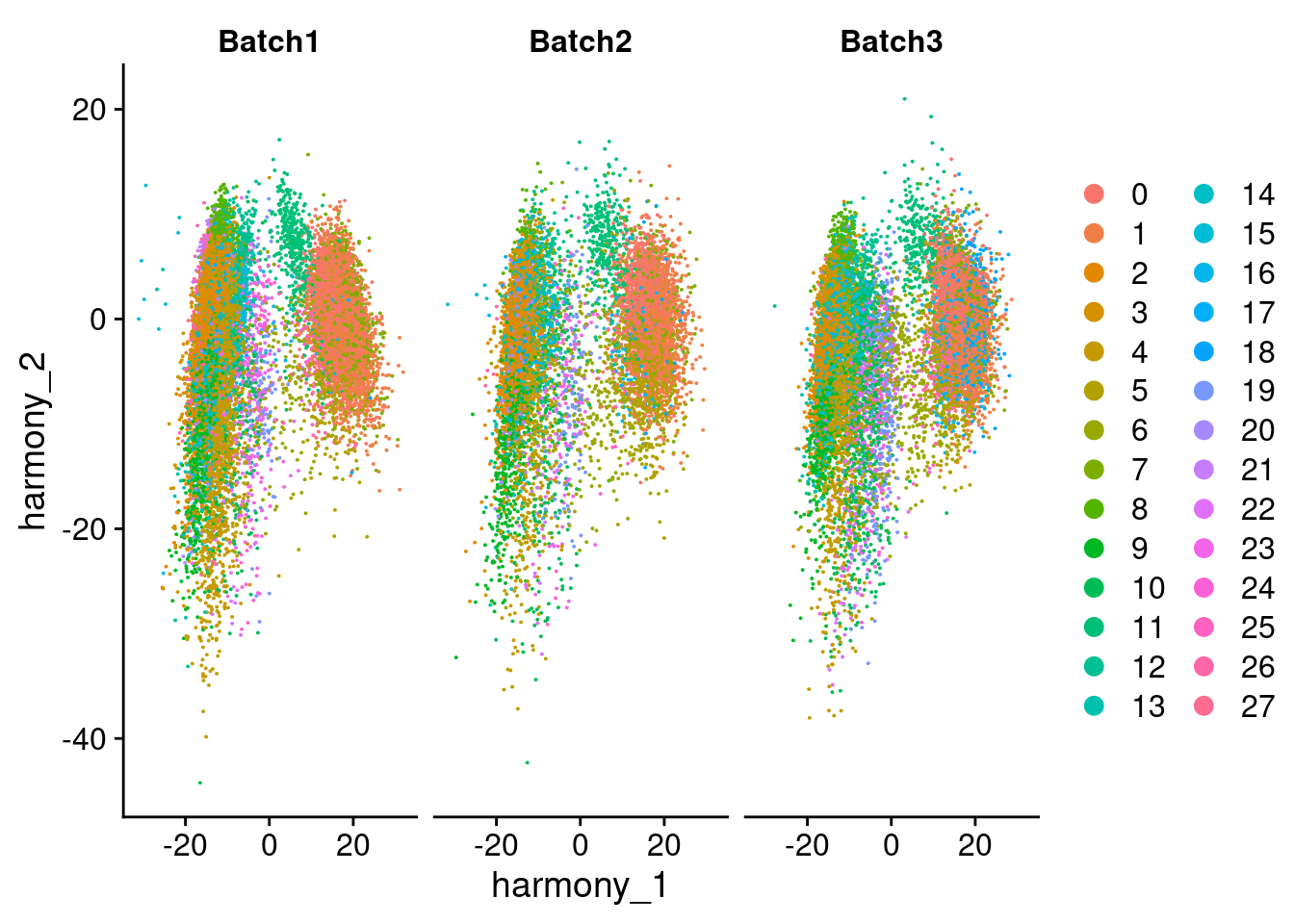
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
VlnPlot(merged, features = c("POU5F1", "PAX6", "TNNT2", "SOX17", "HAND1", "LUM"), ncol=2)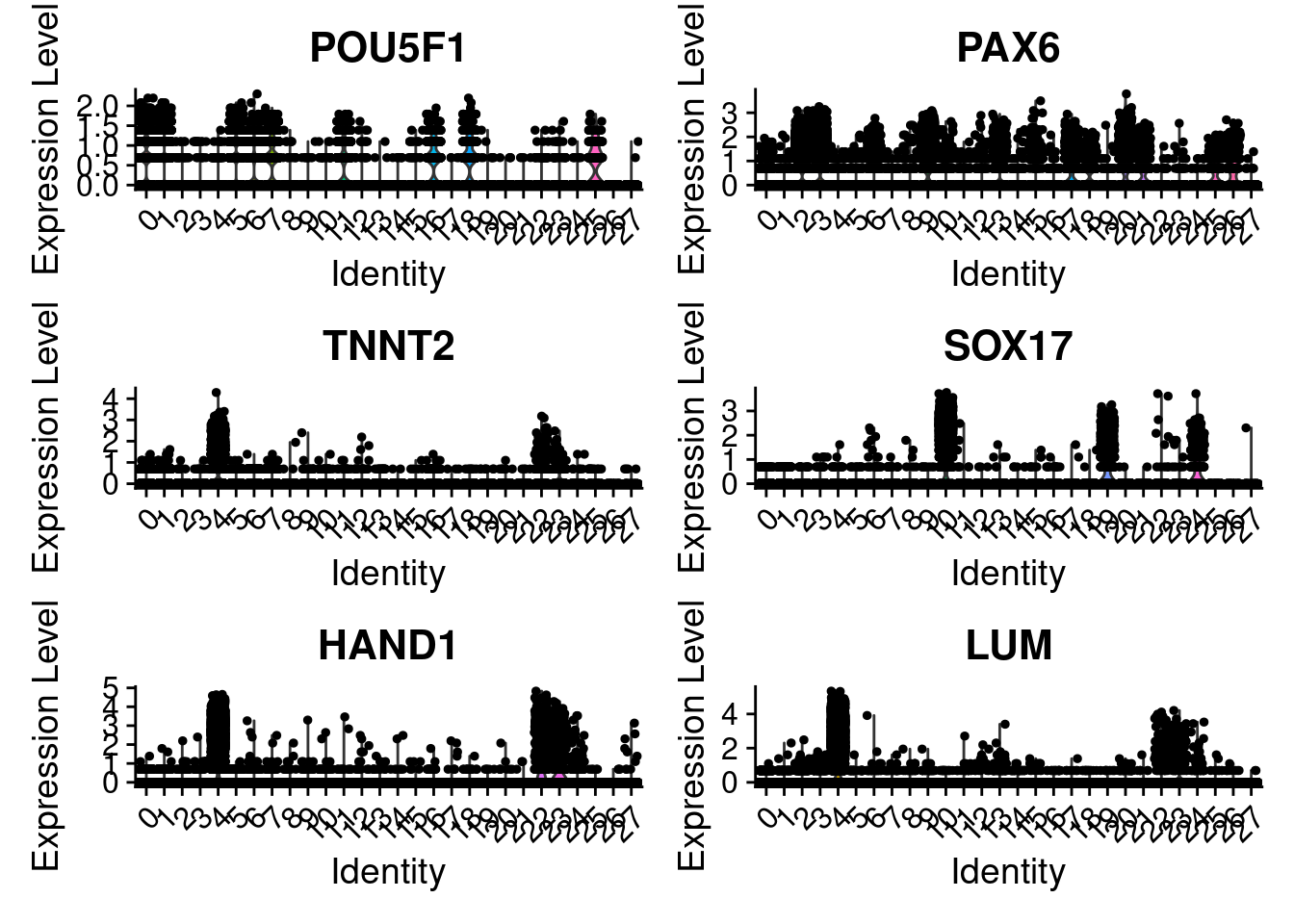
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
#pluripotent markers
FeaturePlot(merged, features = c("POU5F1", "SOX2", "NANOG"), pt.size = 0.2, ncol=3)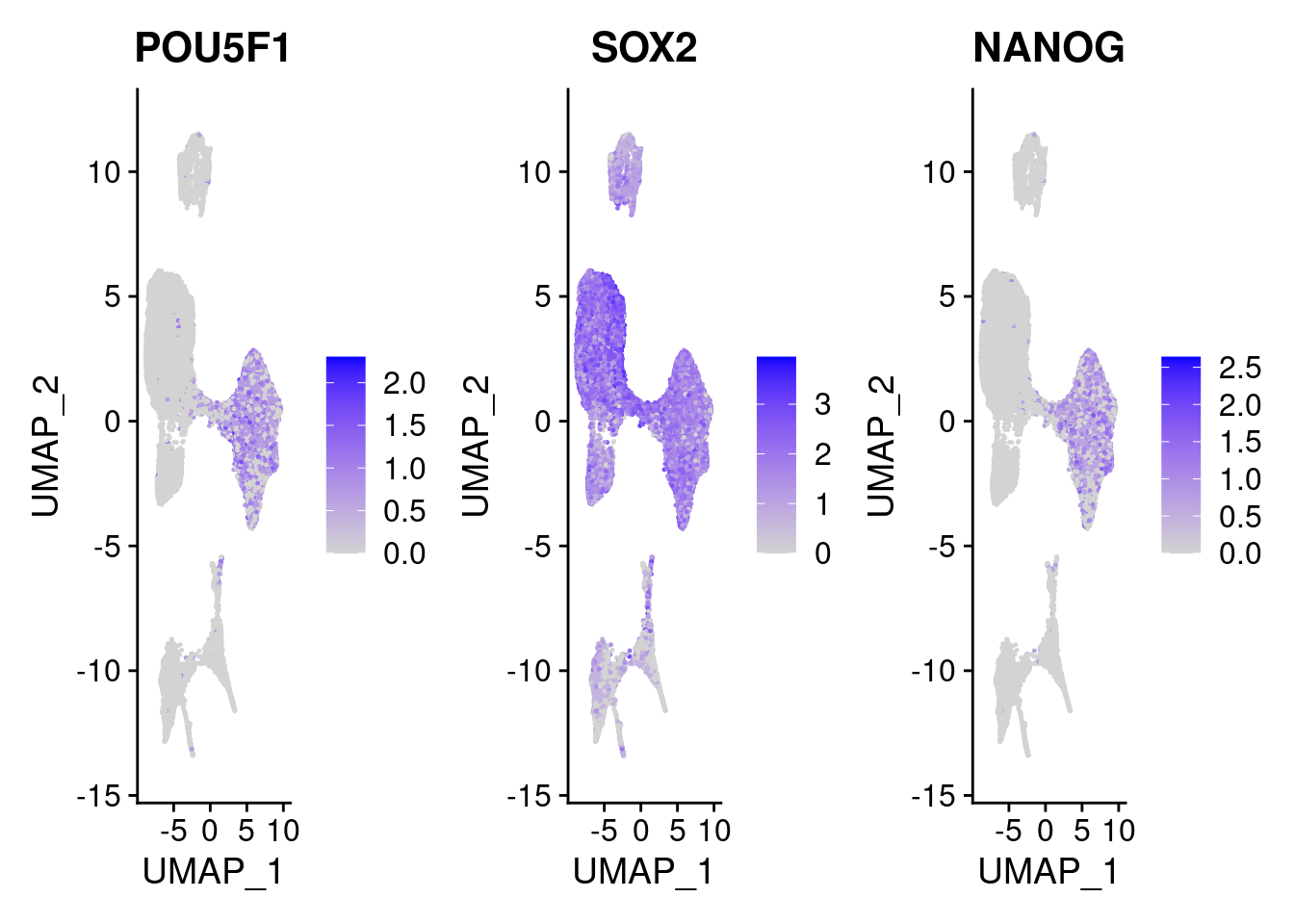
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
#Endoderm markers (first 3 definitive endo, 4-6 liver markers, )
FeaturePlot(merged, features = c("SOX17","CLDN6","FOXA2", "TTR", "AFP", "FGB"), pt.size = 0.2, combine = F)[[1]]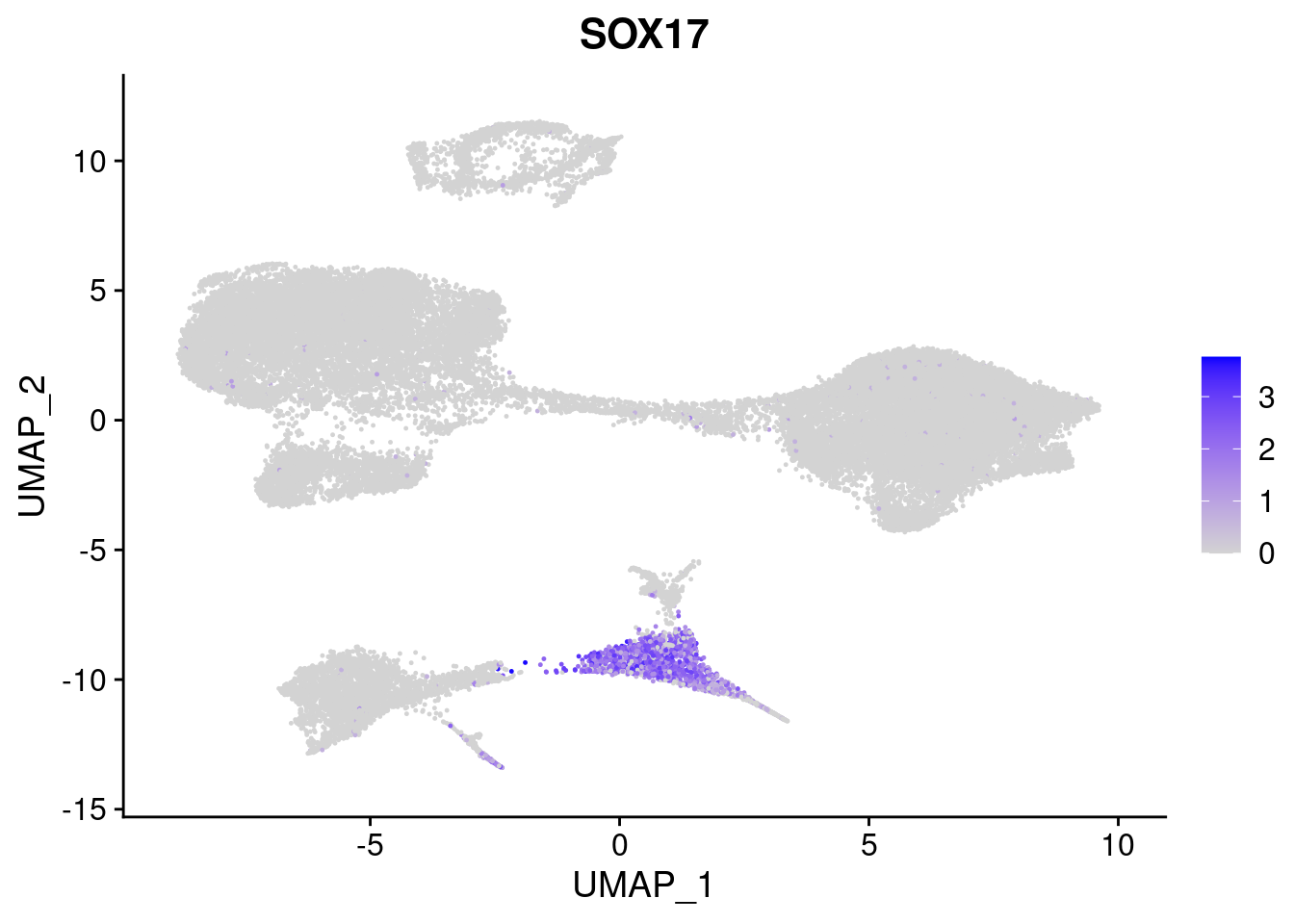
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
[[2]]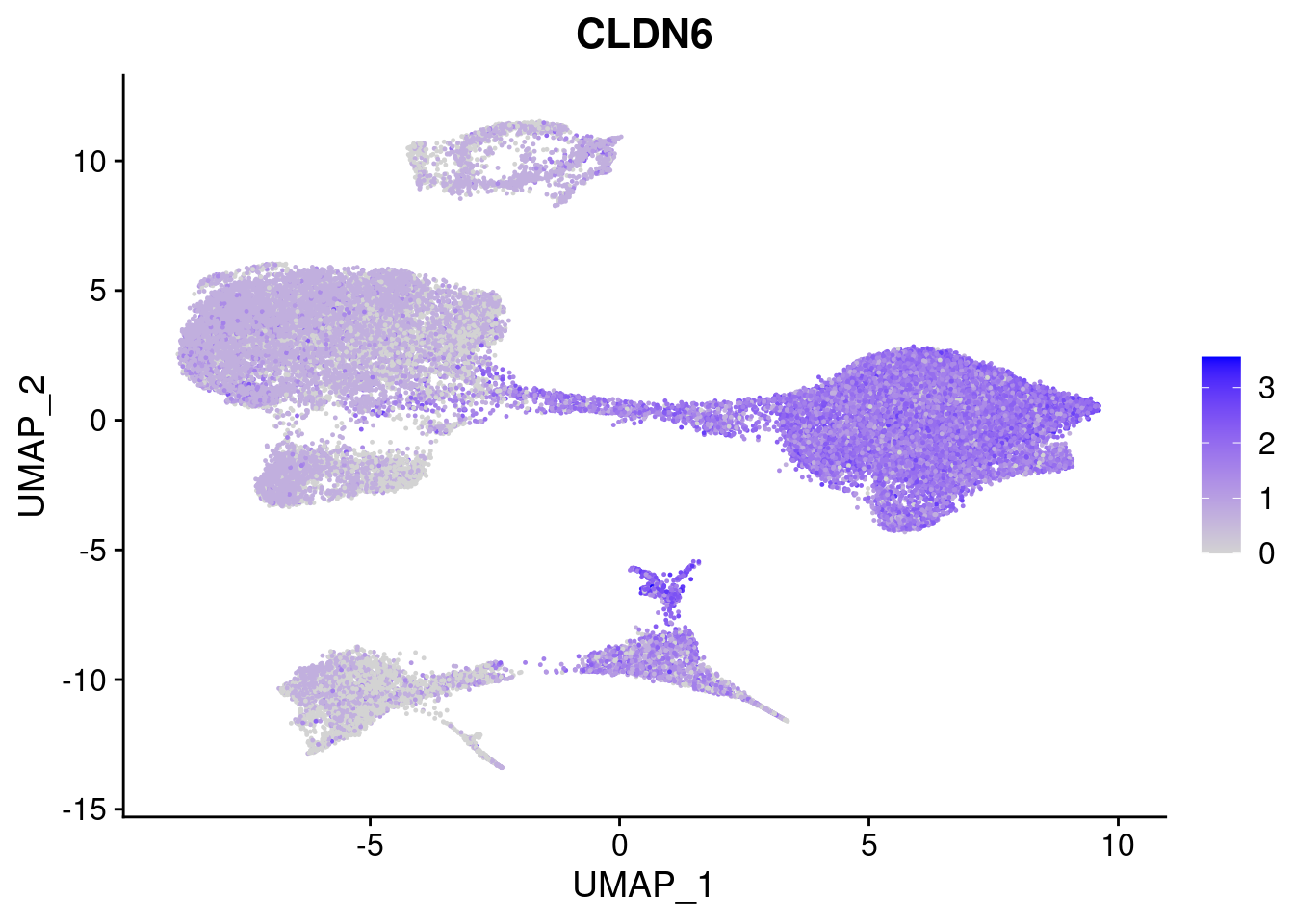
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
[[3]]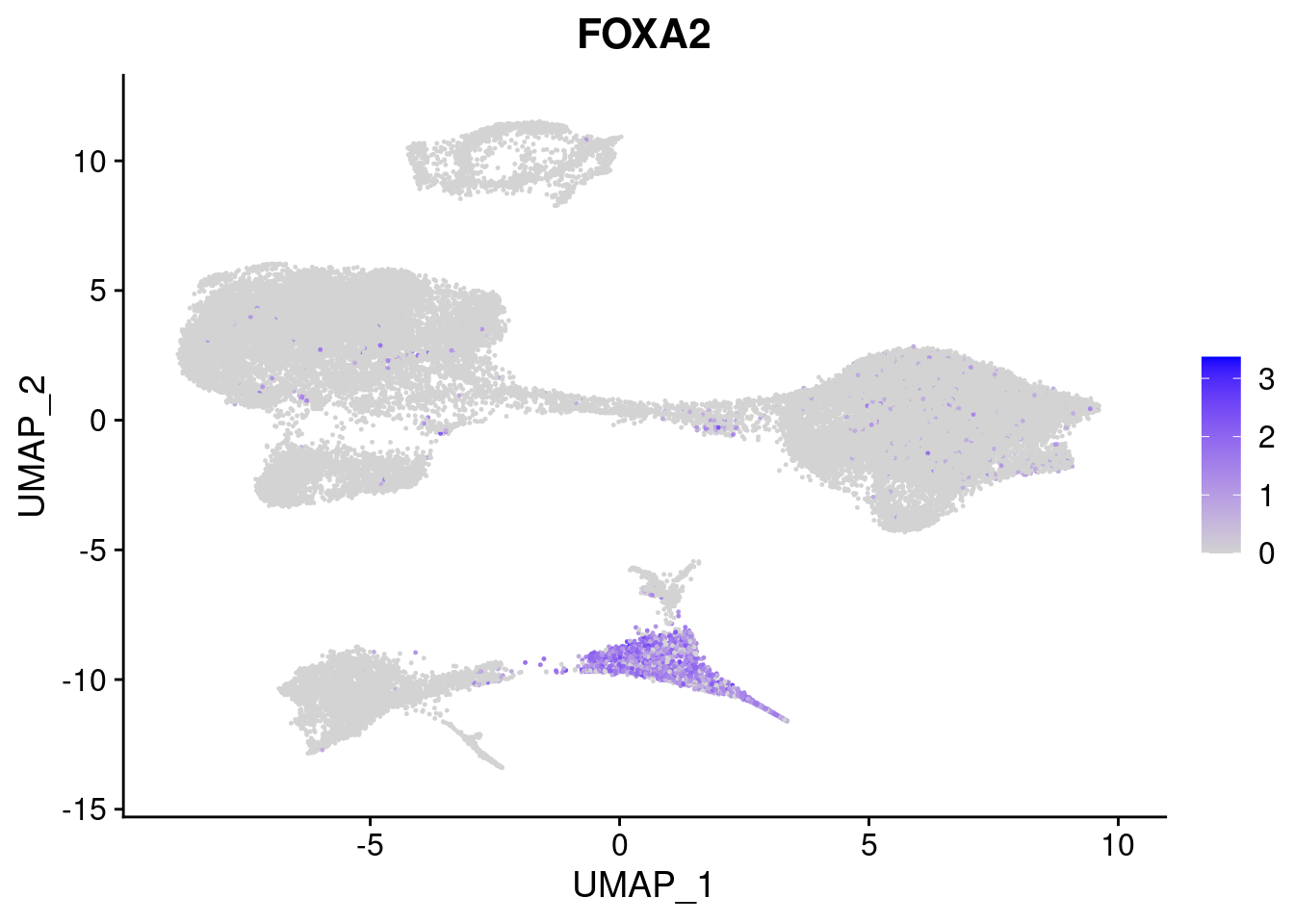
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
[[4]]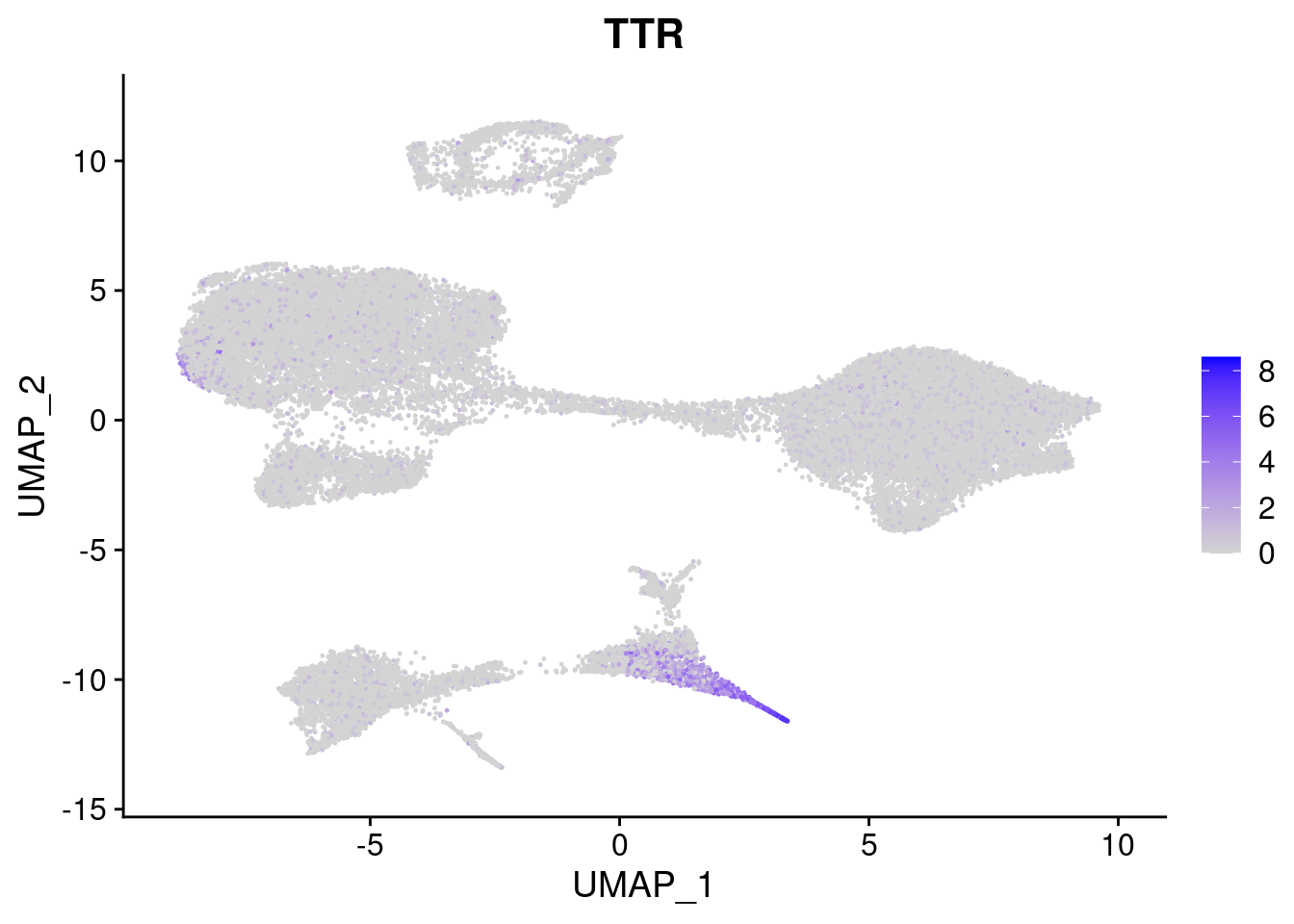
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
[[5]]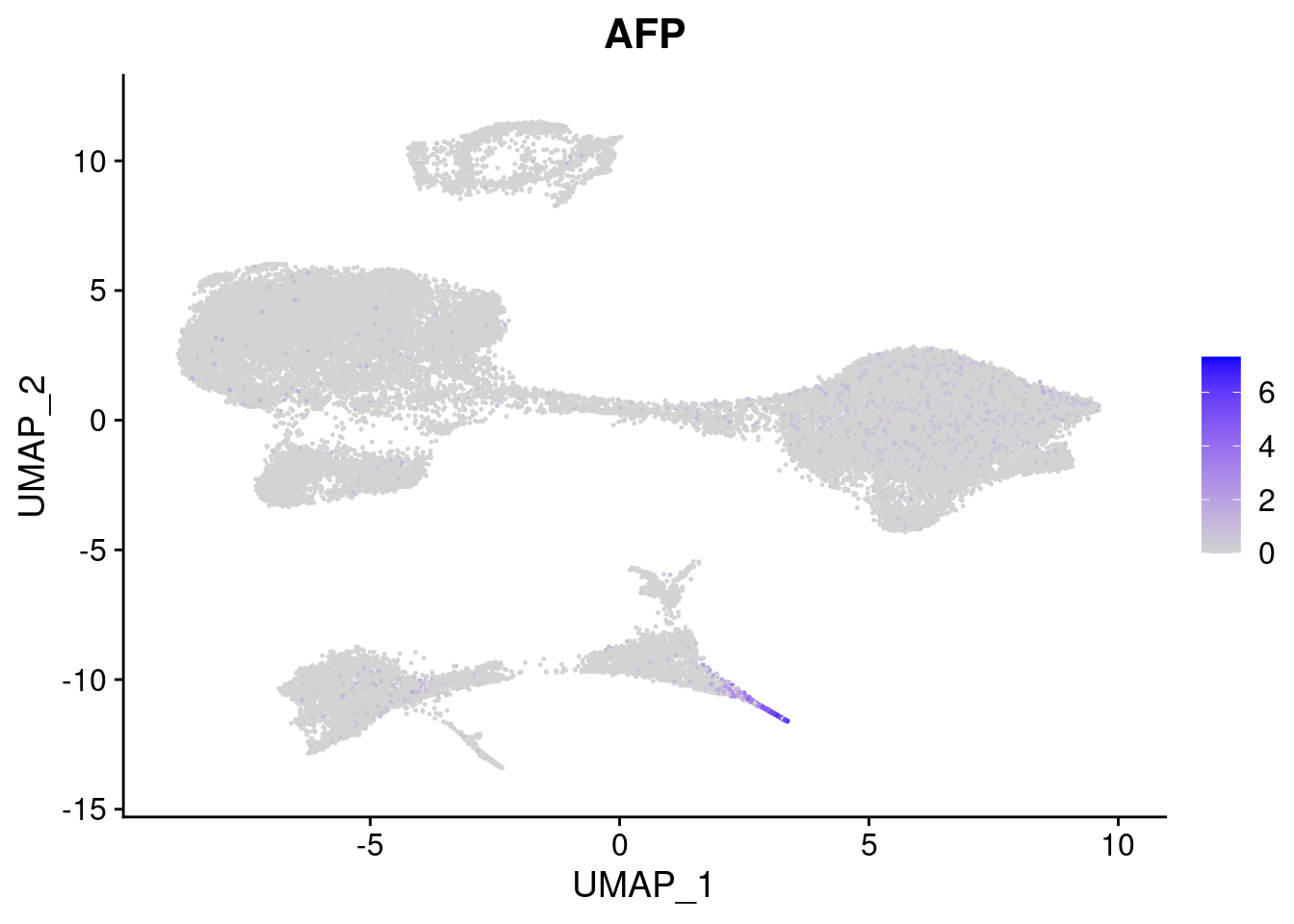
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
[[6]]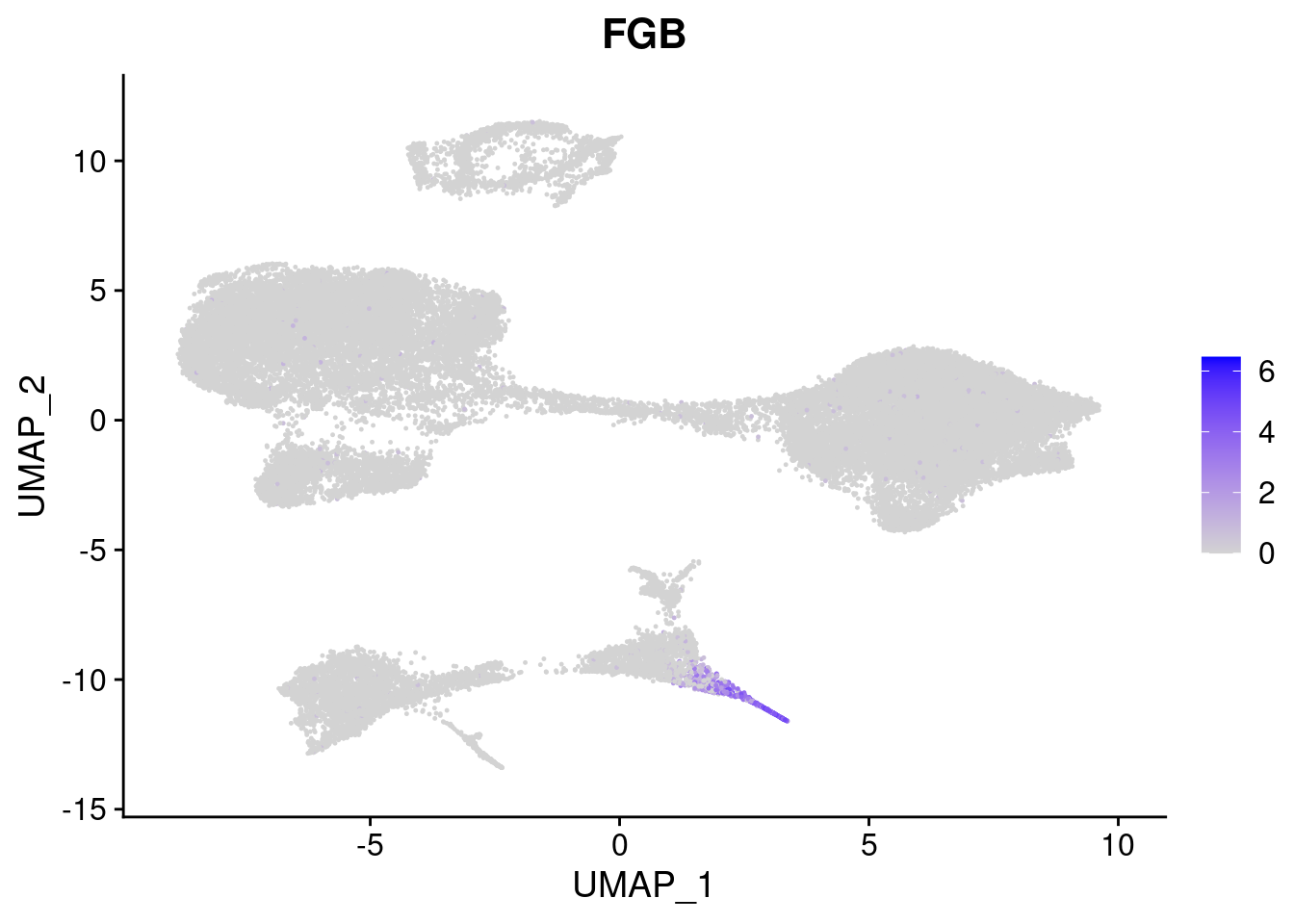
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
#Mesoderm Markers (first 3 early meso markers, 4-6 heart markers, 7-9 endothelial markers (which comes from mesoderm), then some other general muscle markers)
FeaturePlot(merged, features = c("HAND1", "BMP4", "TNNT2","KDR", "GNG11", "ECSCR", "COL3A1", "ACTC1"), pt.size = 0.2, combine=F)[[1]]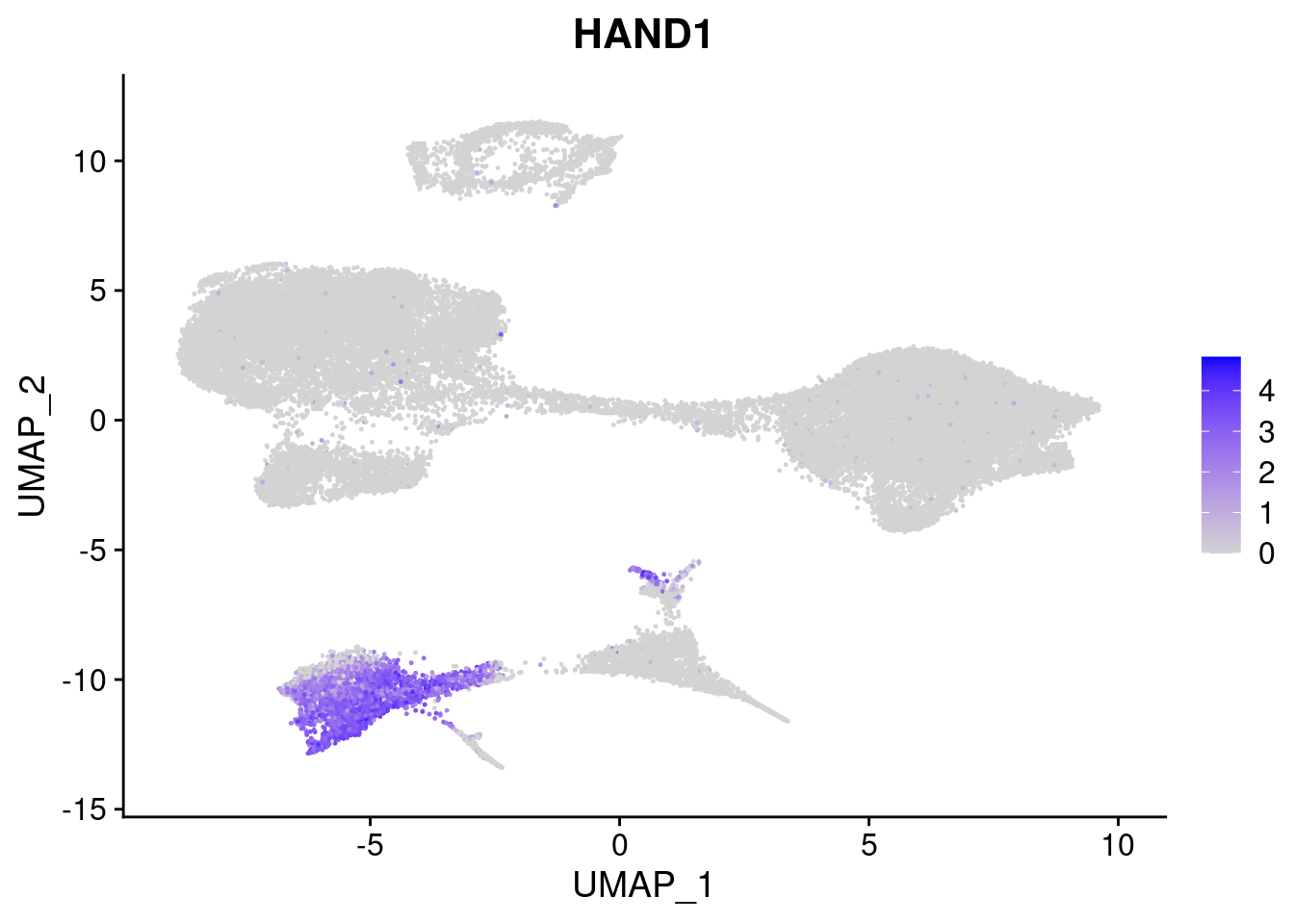
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
[[2]]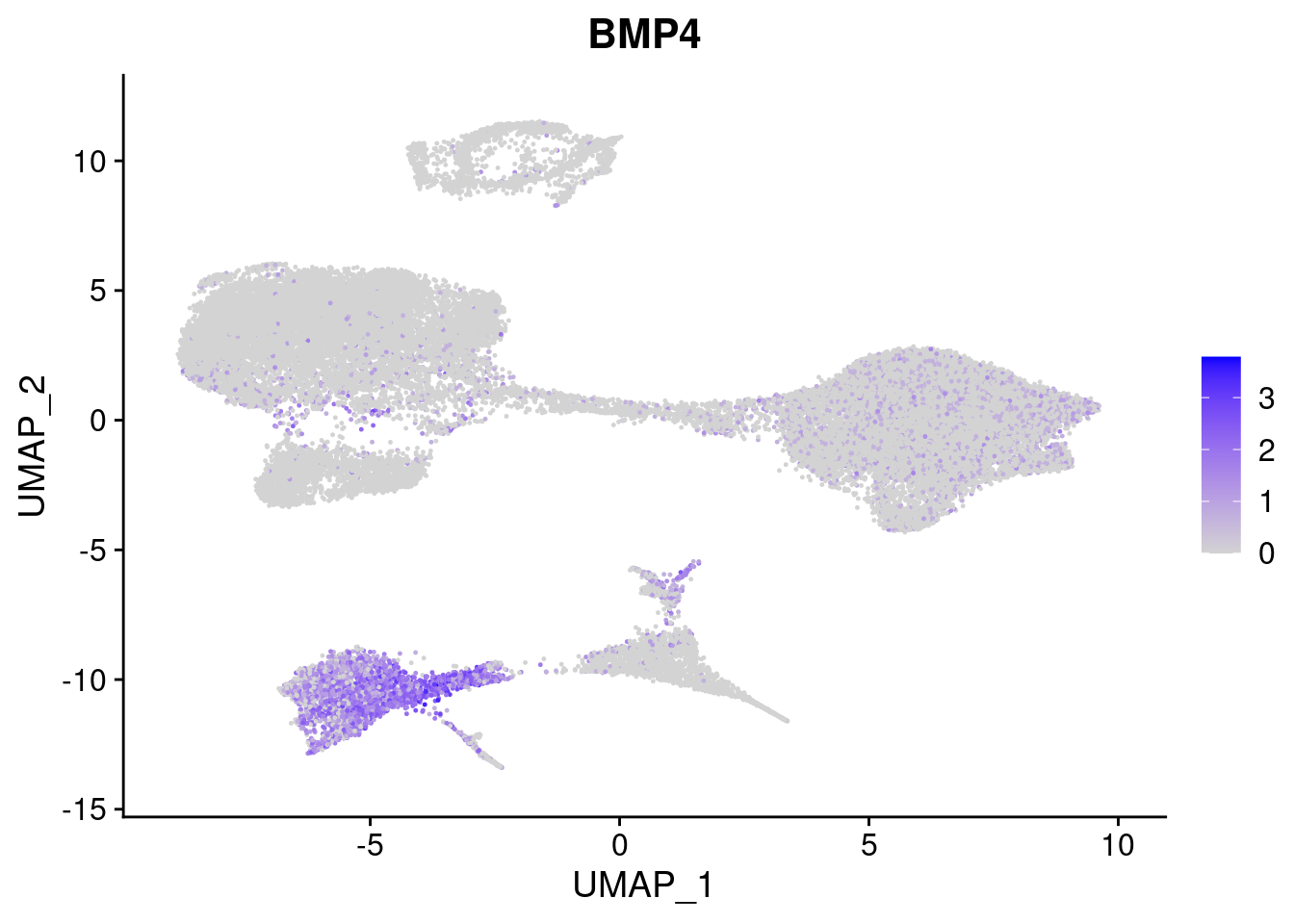
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
[[3]]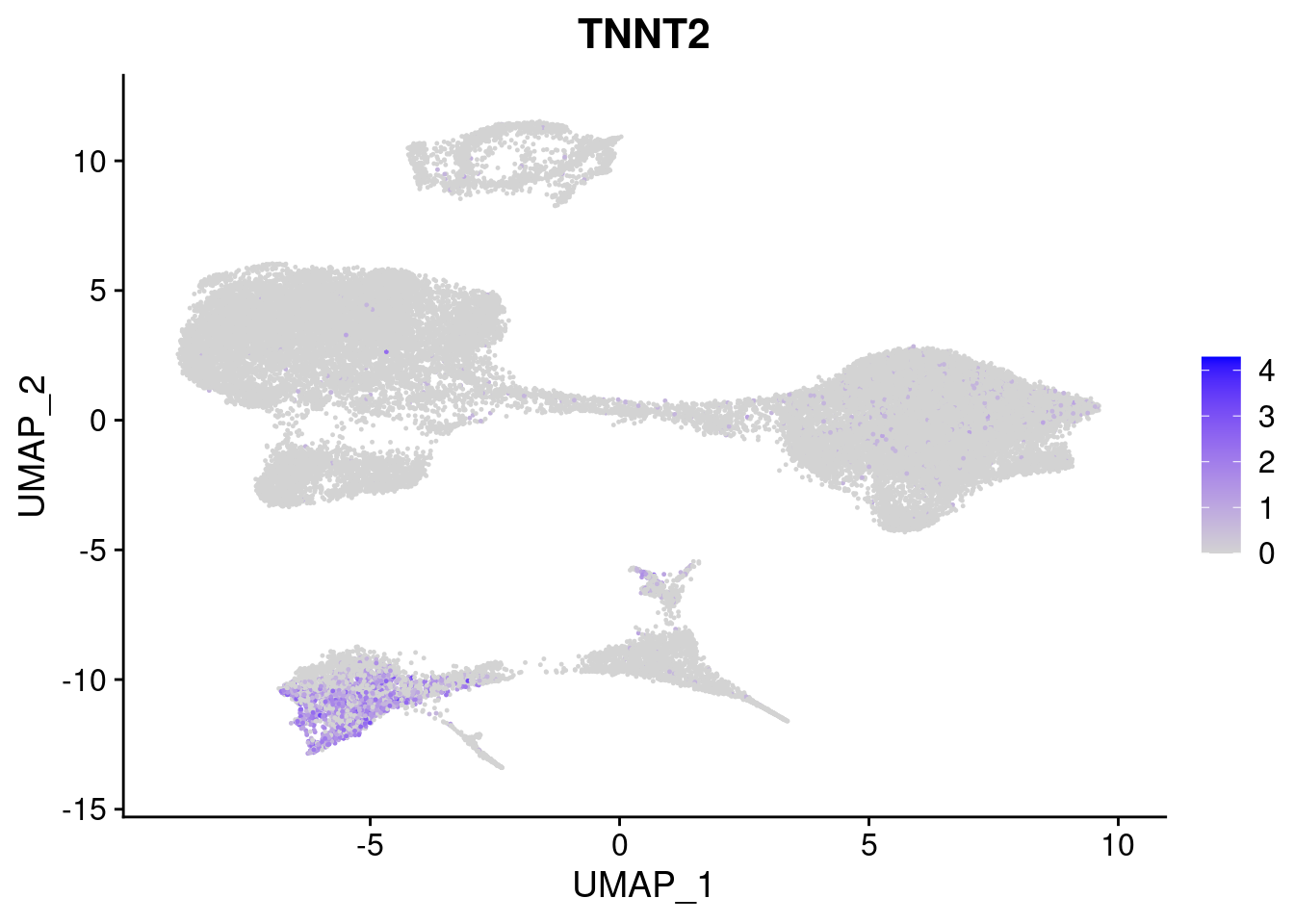
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
[[4]]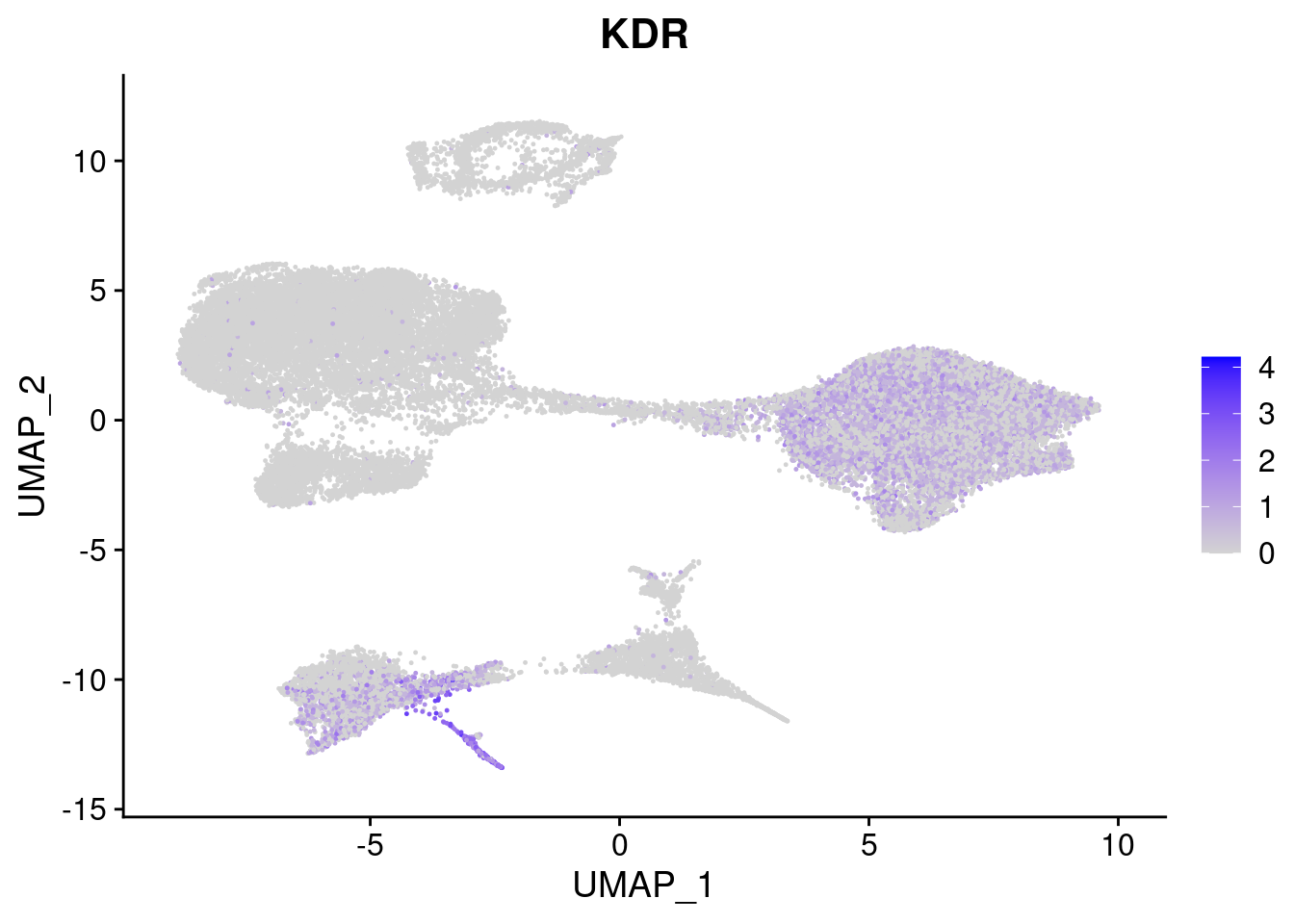
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
[[5]]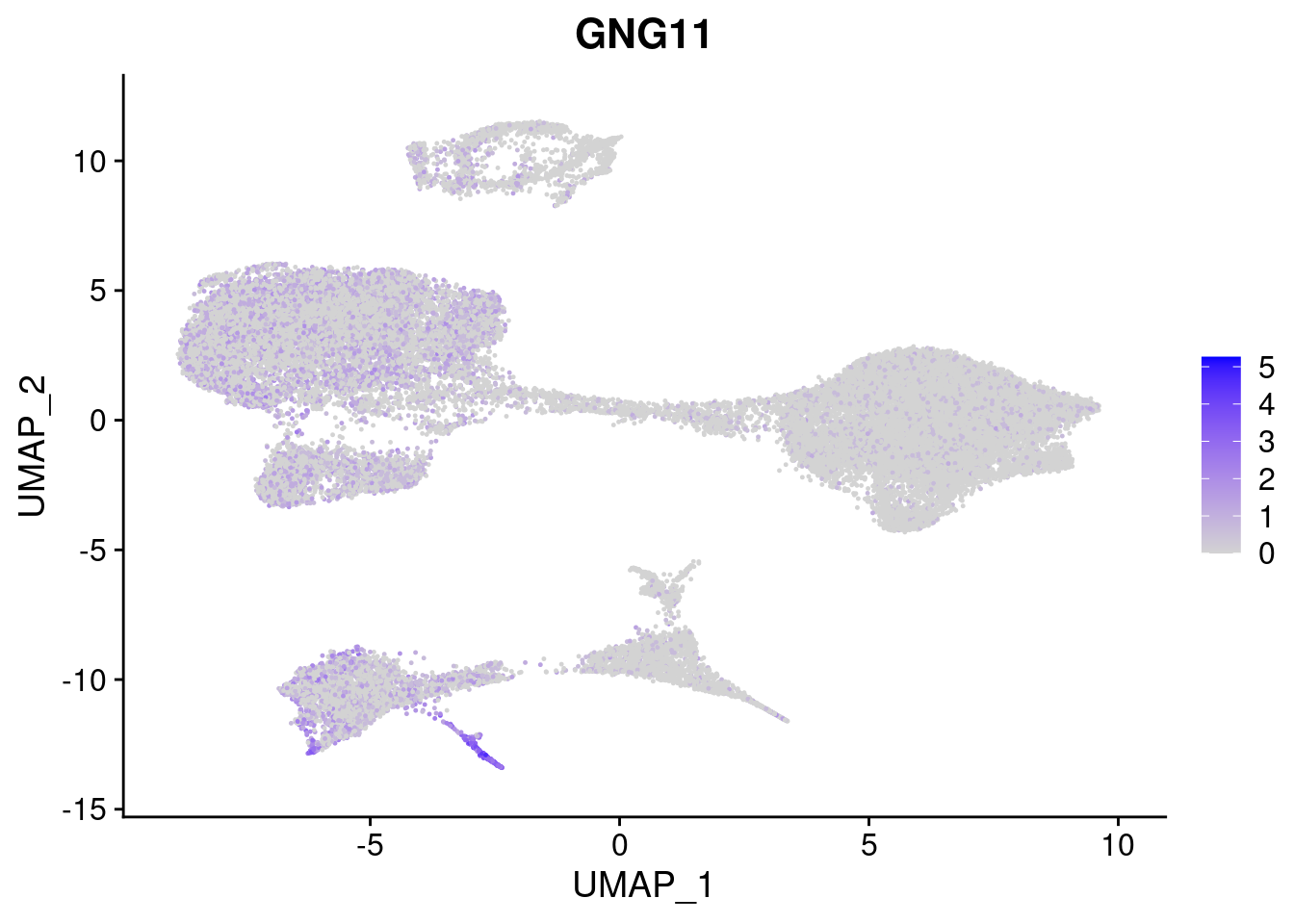
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
[[6]]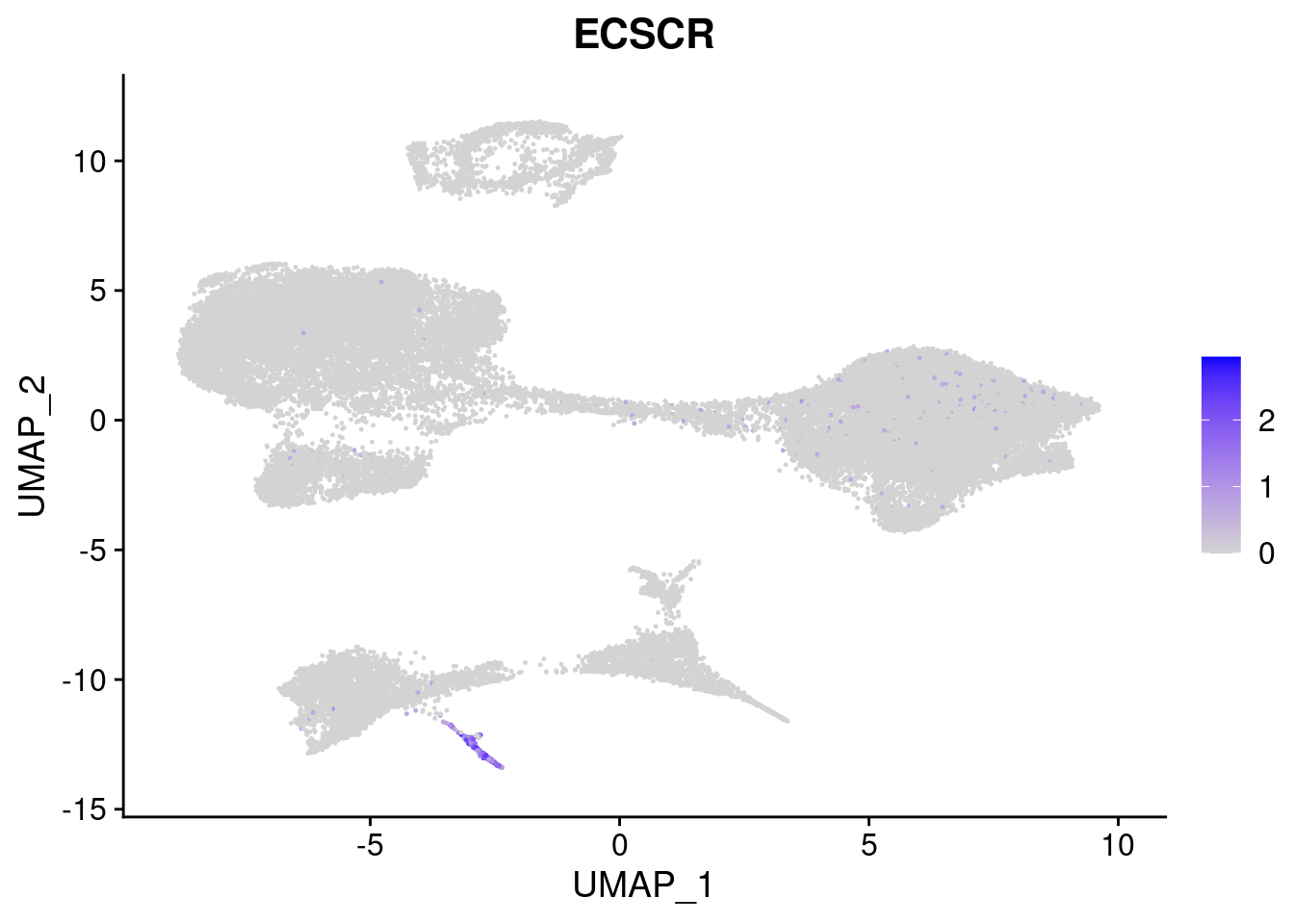
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
[[7]]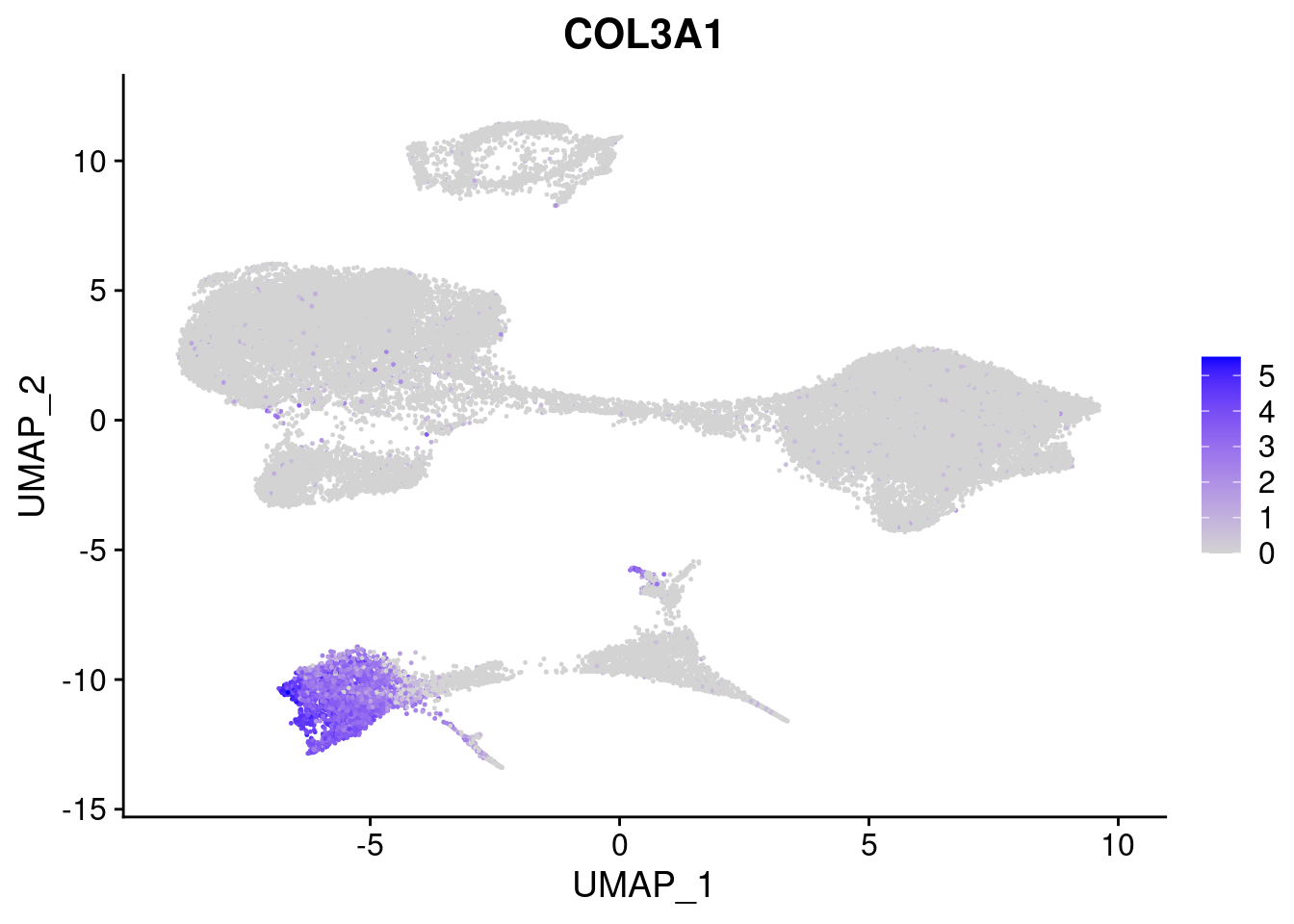
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
[[8]]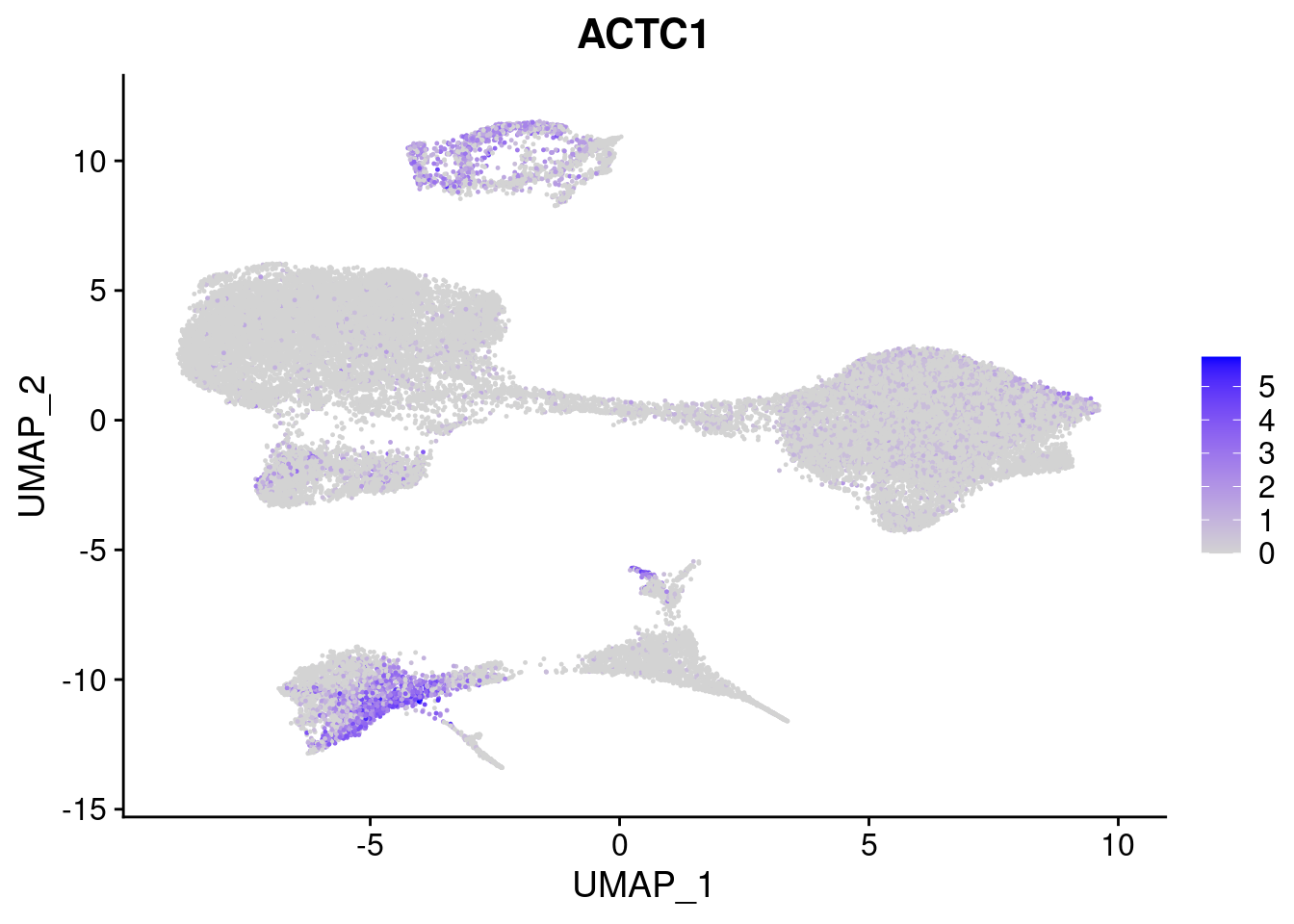
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
#Ectoderm Markers (3-1 early ectoderm markers, 4-6schwann cell (myelinating, non myelinating, or precursor), 7-8 oligodendrocytes, 9-10 radial glia)
FeaturePlot(merged, features = c("PAX6", "GBX2", "NES", "MPZ", "SOX10","GAP43", "OLIG1", "OLIG2", "VIM", "HES5"), pt.size = 0.2, ncol=3, combine=F)[[1]]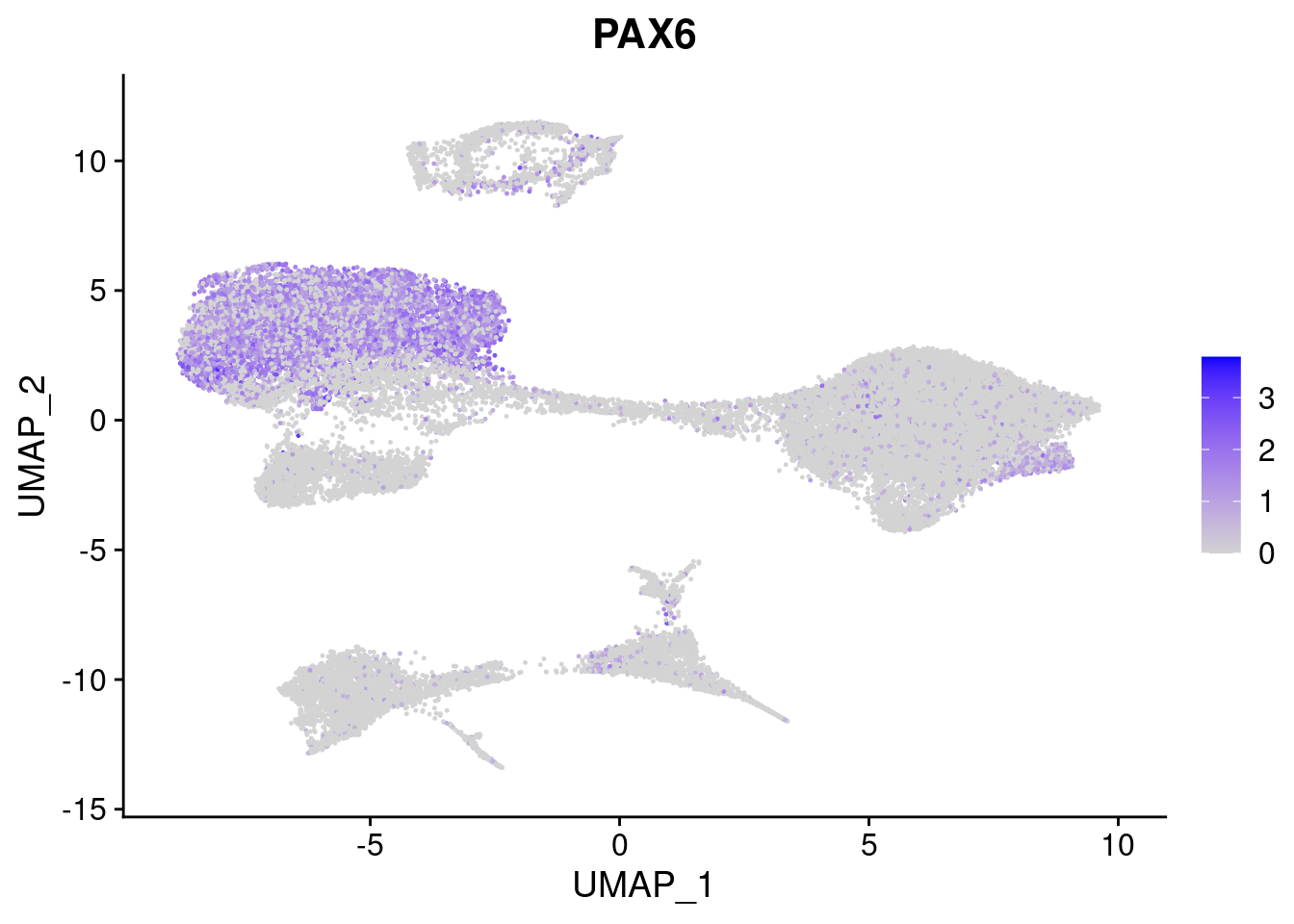
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
[[2]]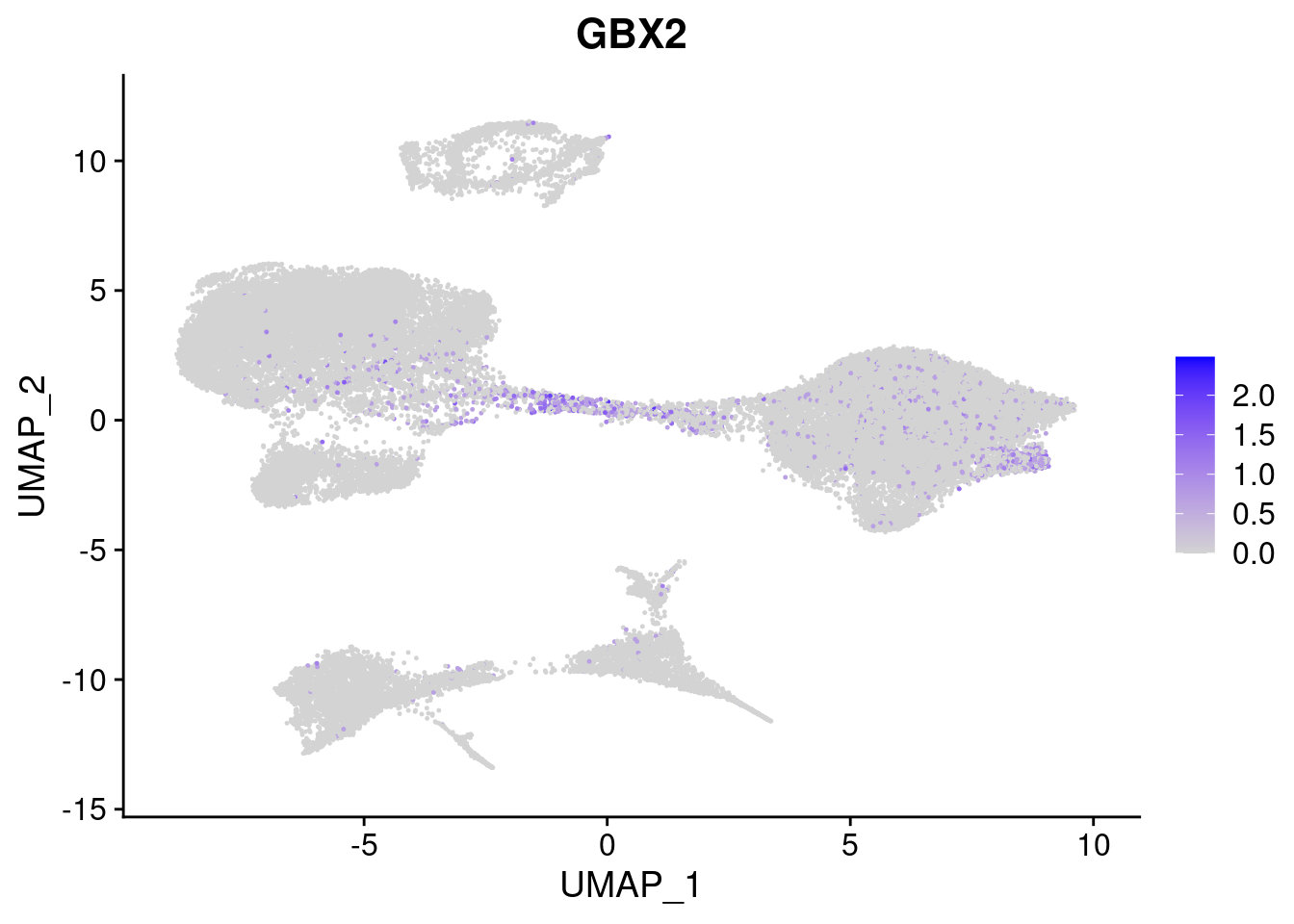
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
[[3]]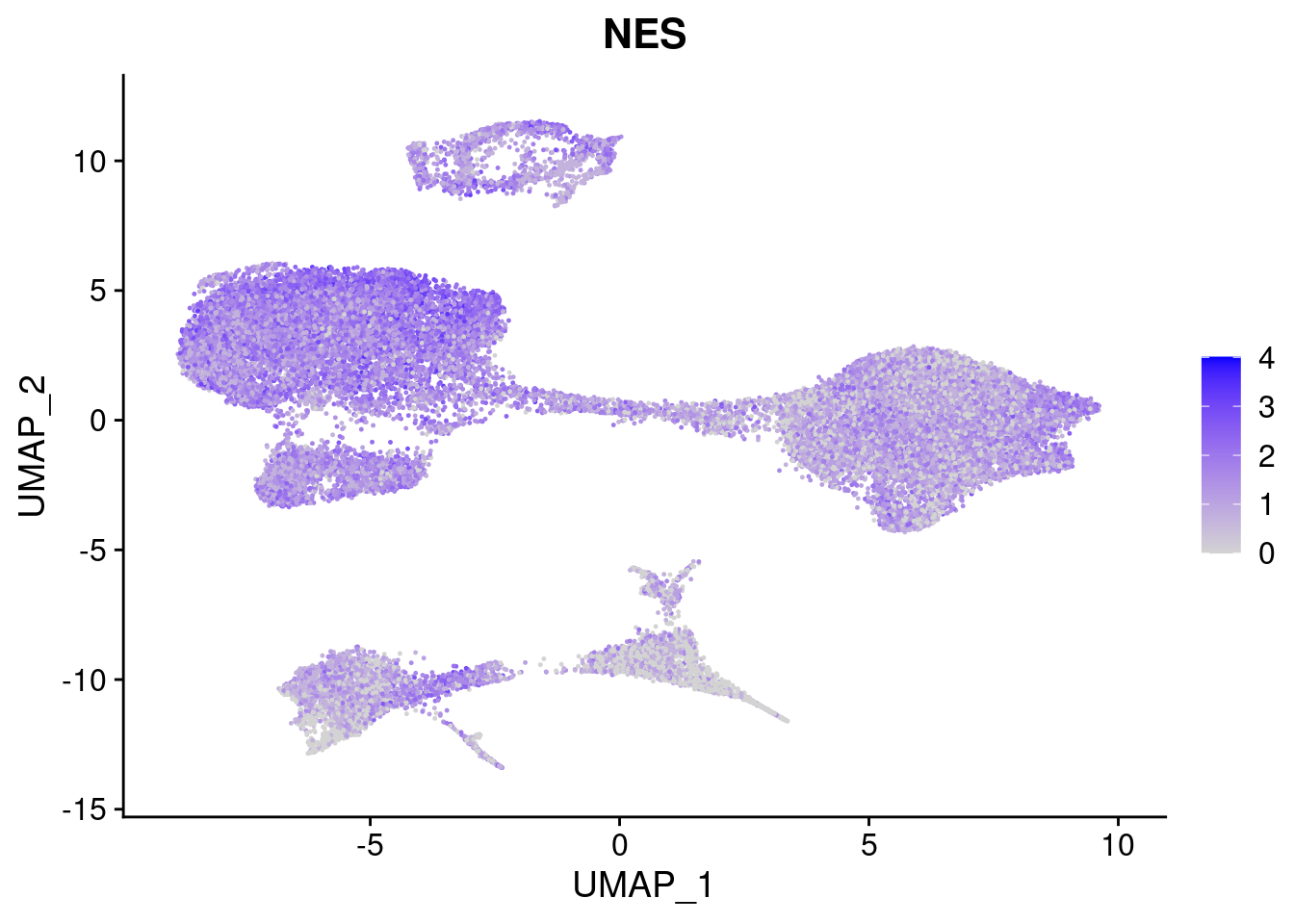
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
[[4]]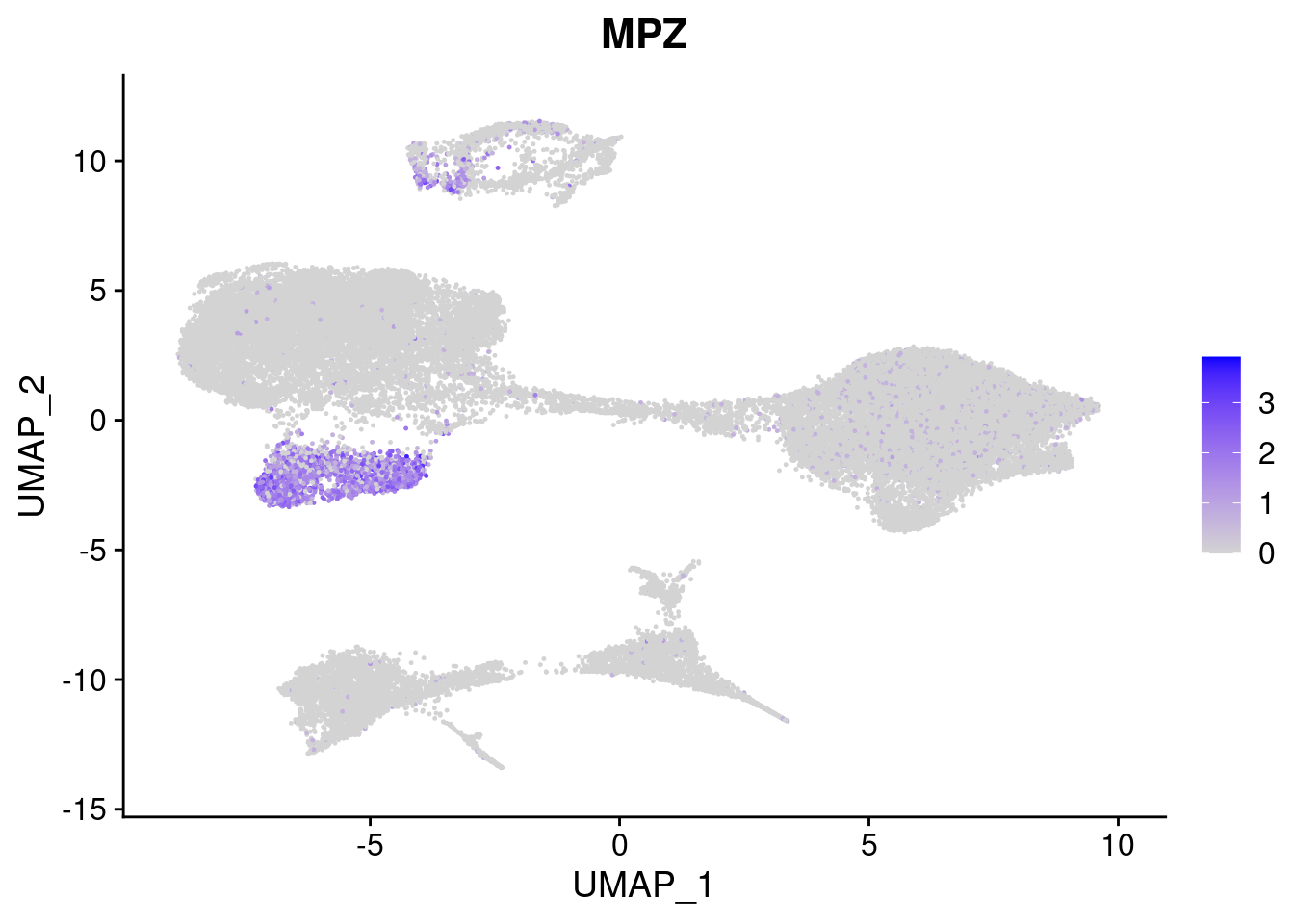
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
[[5]]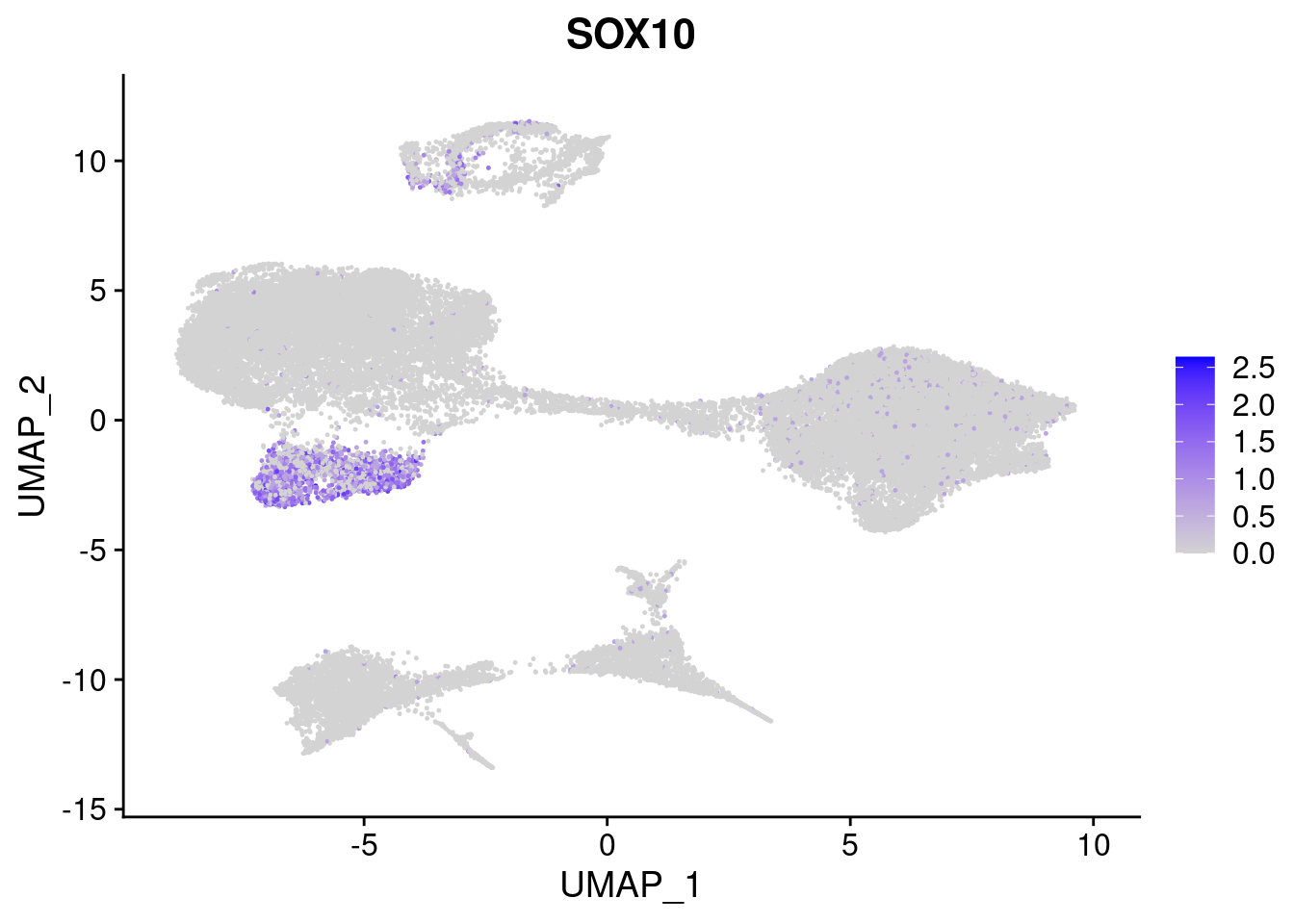
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
[[6]]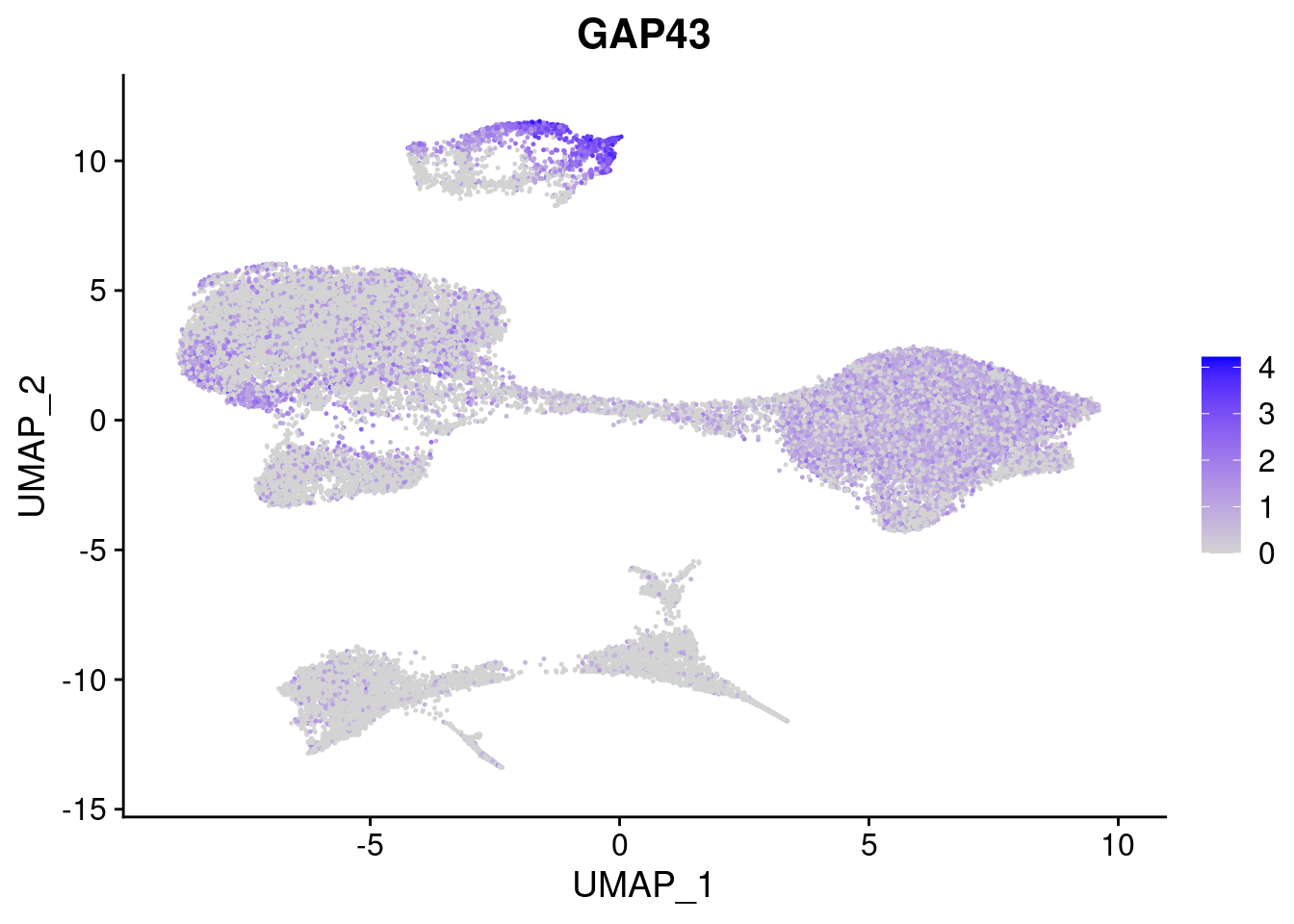
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
[[7]]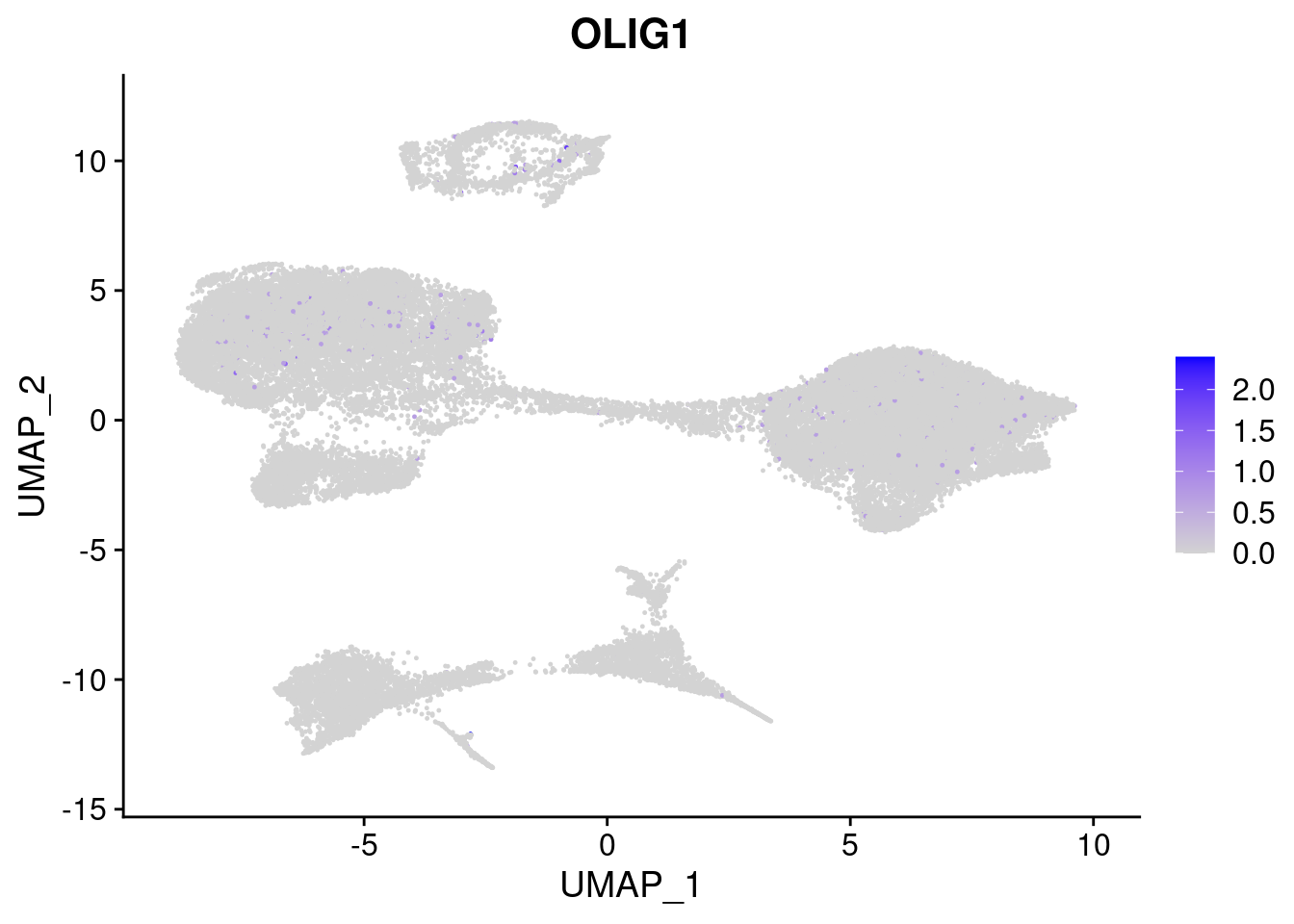
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
[[8]]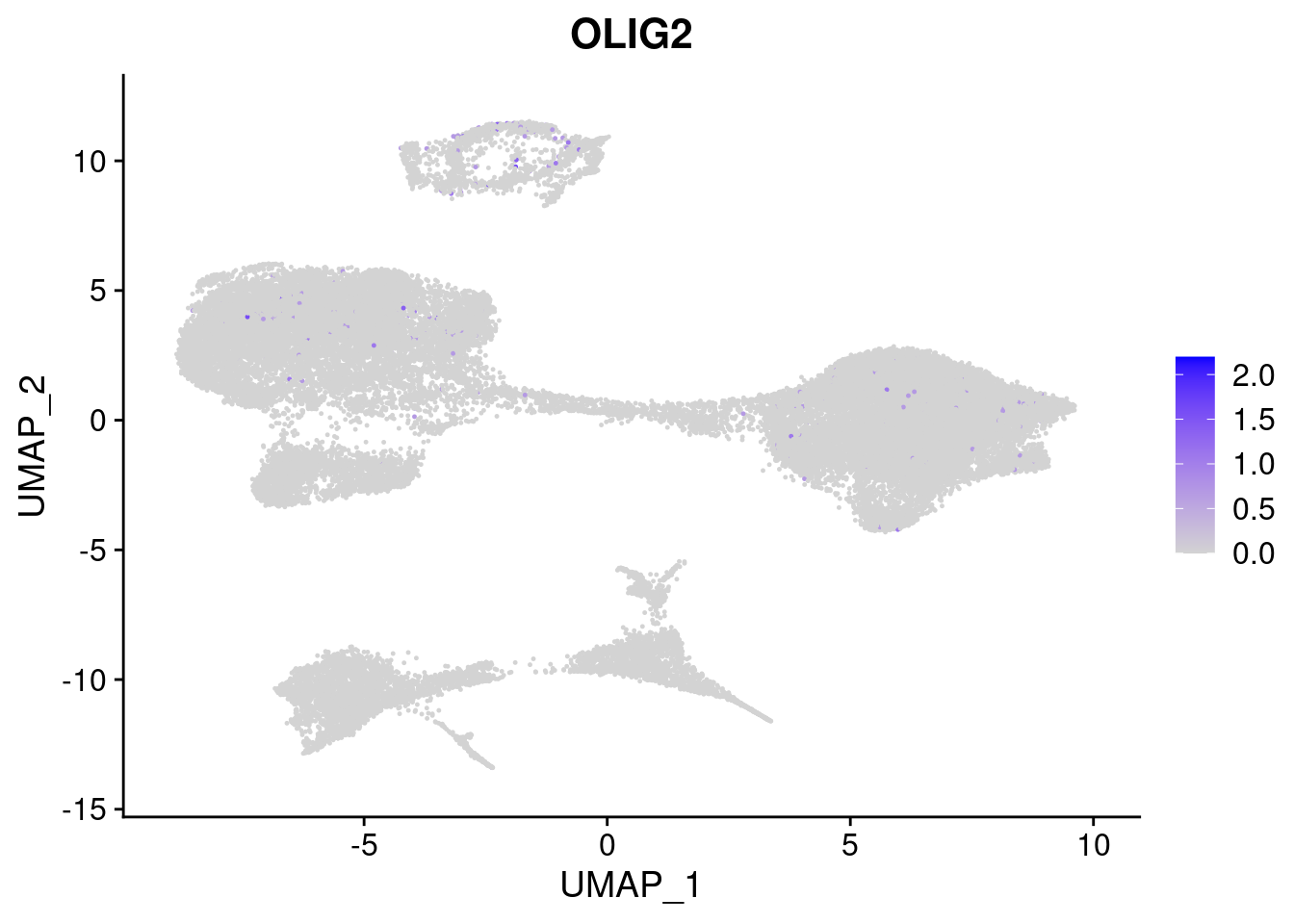
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
[[9]]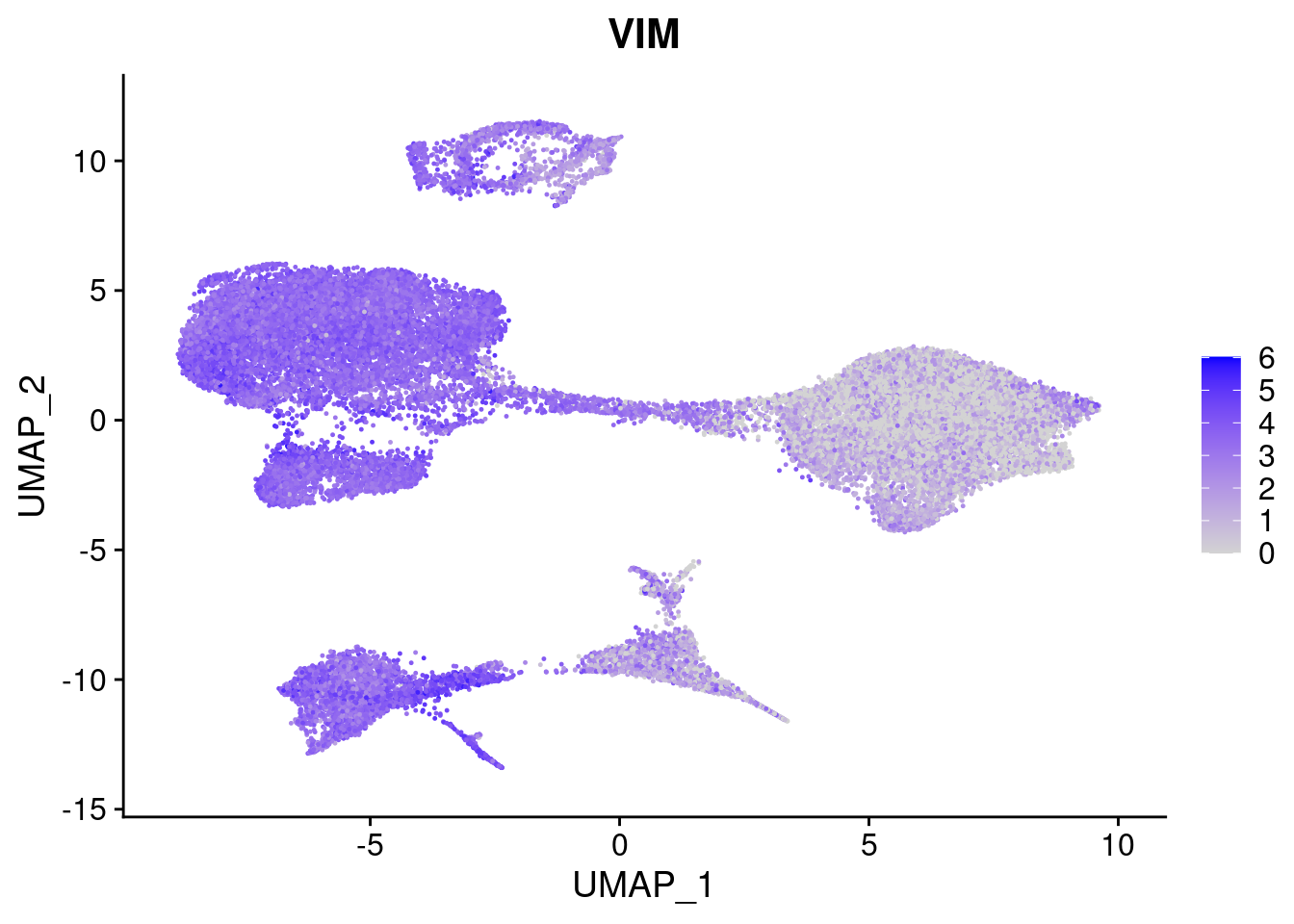
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
[[10]]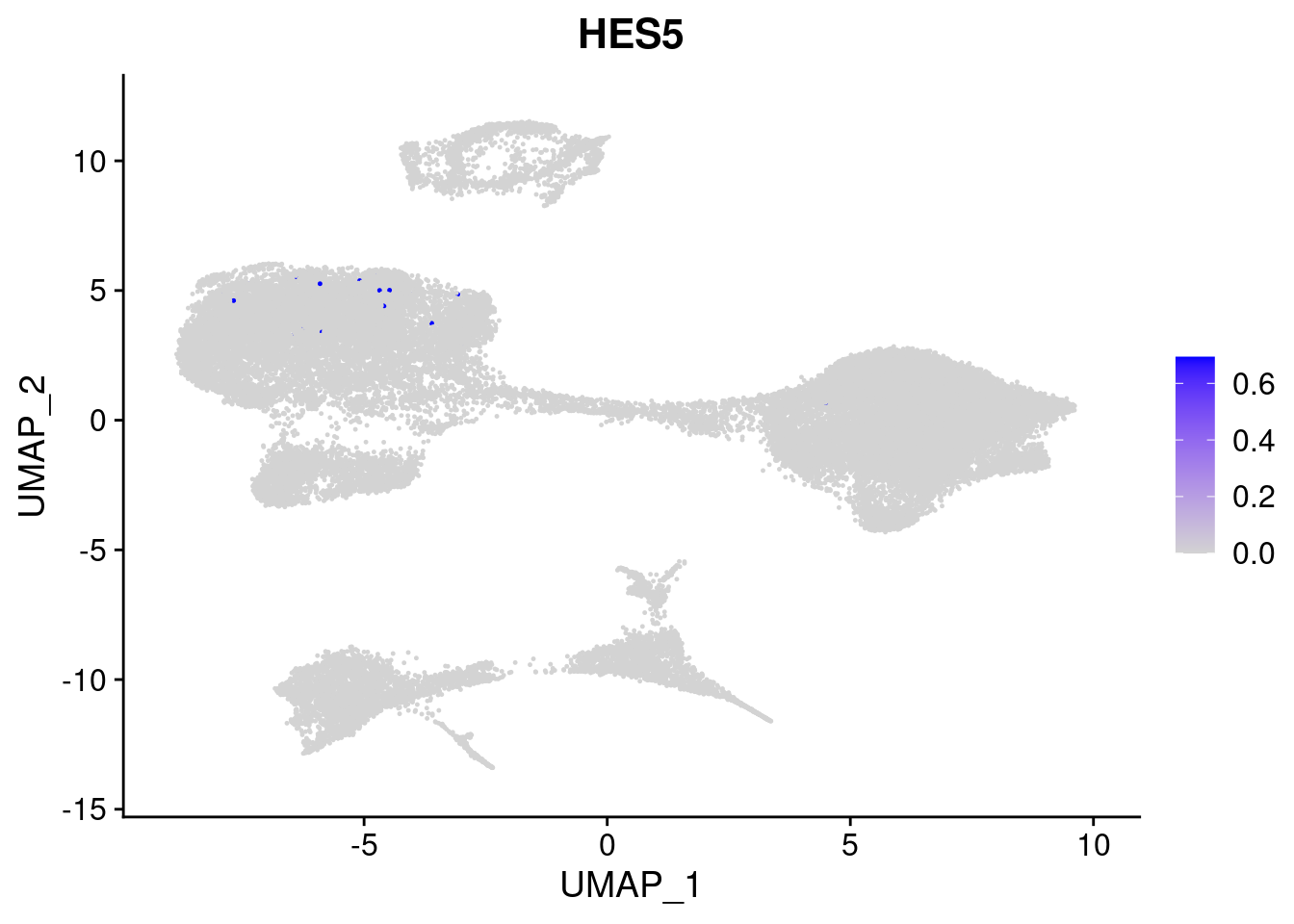
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
#More ectoderm, specifically neurons
#immature neurons: NEUROD1
#Mature Neurons: MAP2, SYP
#dopaminergic: TH, FOXA2,
FeaturePlot(merged, features = c("MAP2", "SYP","NEUROD1", "TH" ), pt.size = 0.2, ncol=3)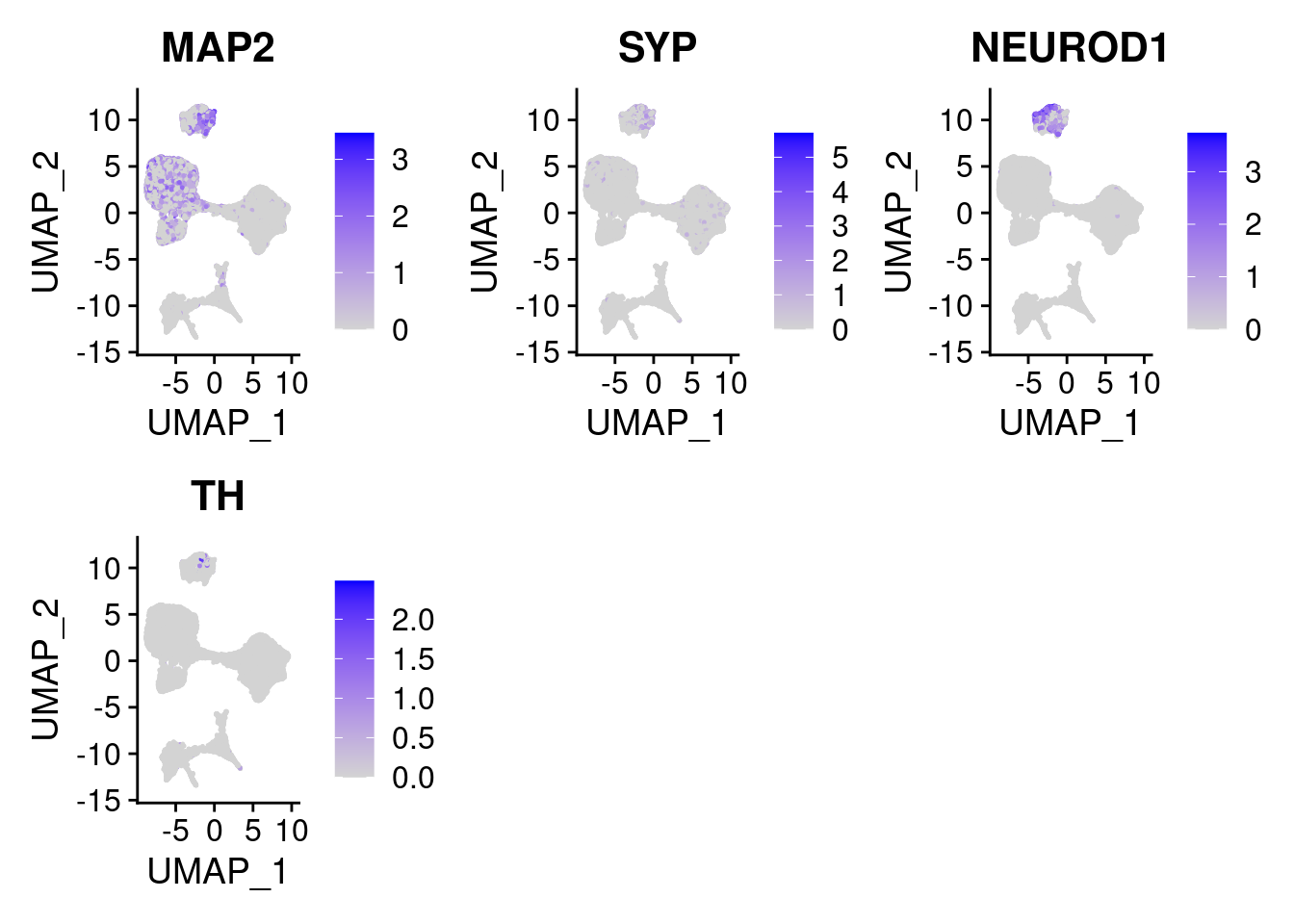
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
Identify cluster markers
#how many cells per cluster?
t1<-table(merged@meta.data$SCT_snn_res.1, merged@meta.data$Batch)
t1
Batch1 Batch2 Batch3
0 2377 1416 2350
1 2128 1191 1821
2 2014 1306 1157
3 1708 721 878
4 1361 435 871
5 406 855 741
6 412 512 1075
7 687 258 647
8 778 221 526
9 550 490 442
10 249 273 859
11 445 410 309
12 615 207 329
13 416 141 540
14 443 190 404
15 332 203 320
16 36 92 592
17 256 245 207
18 11 55 633
19 105 120 369
20 289 255 43
21 568 3 0
22 82 75 252
23 238 64 94
24 103 60 134
25 83 38 99
26 101 19 29
27 80 10 29#how many cells per cluster from each individual?
t2<-table(merged@meta.data$SCT_snn_res.1, merged@meta.data$individual)
t2
SNG-NA18511 SNG-NA18858 SNG-NA19160
0 363 5633 147
1 199 4859 82
2 3005 237 1235
3 1575 154 1578
4 831 31 1805
5 113 1741 148
6 871 289 839
7 84 1466 42
8 155 34 1336
9 971 103 408
10 524 38 819
11 106 965 93
12 139 19 993
13 89 11 997
14 135 7 895
15 581 37 237
16 411 16 293
17 416 56 236
18 450 5 244
19 155 15 424
20 133 435 19
21 328 33 210
22 108 6 295
23 83 135 178
24 90 3 204
25 18 193 9
26 0 149 0
27 16 0 103Reclustering with less resolution, check if everything is robust
#reassign idents
Idents(merged)<- 'SCT_snn_res.0.5'DimPlot(merged, reduction = "umap")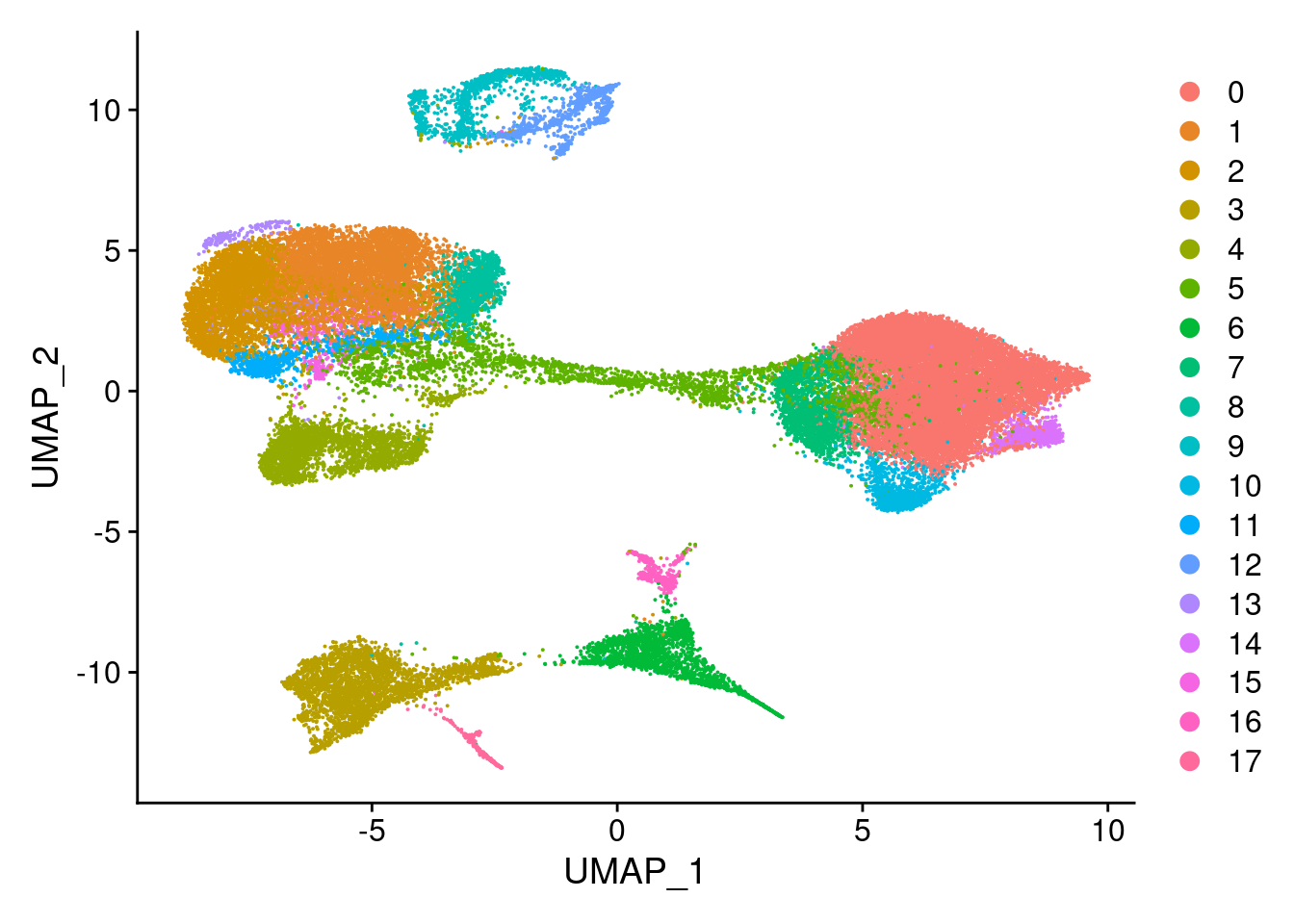
DimPlot(merged, reduction = "umap", group.by = "Batch")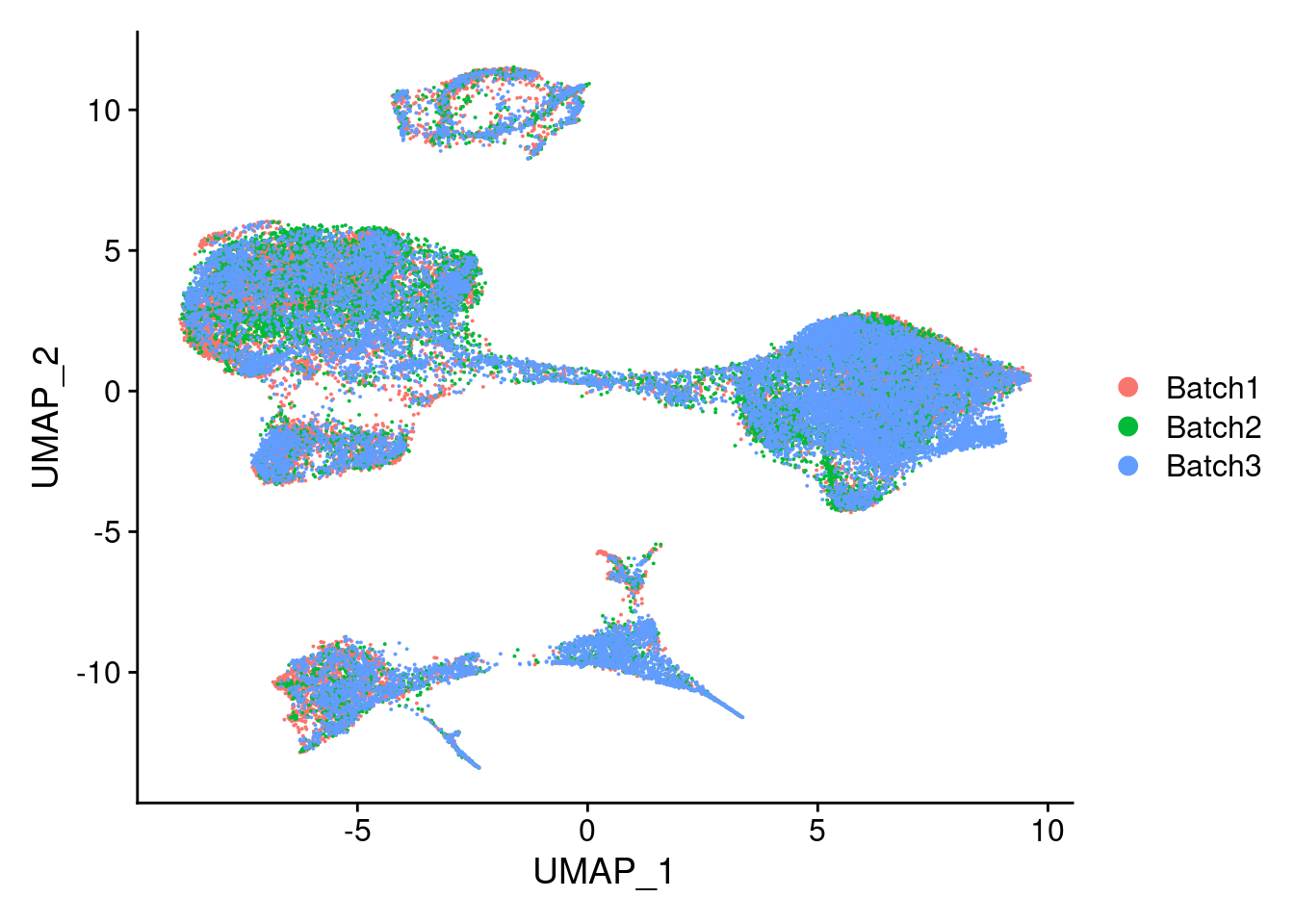
DimPlot(merged, reduction = "umap", group.by = "individual")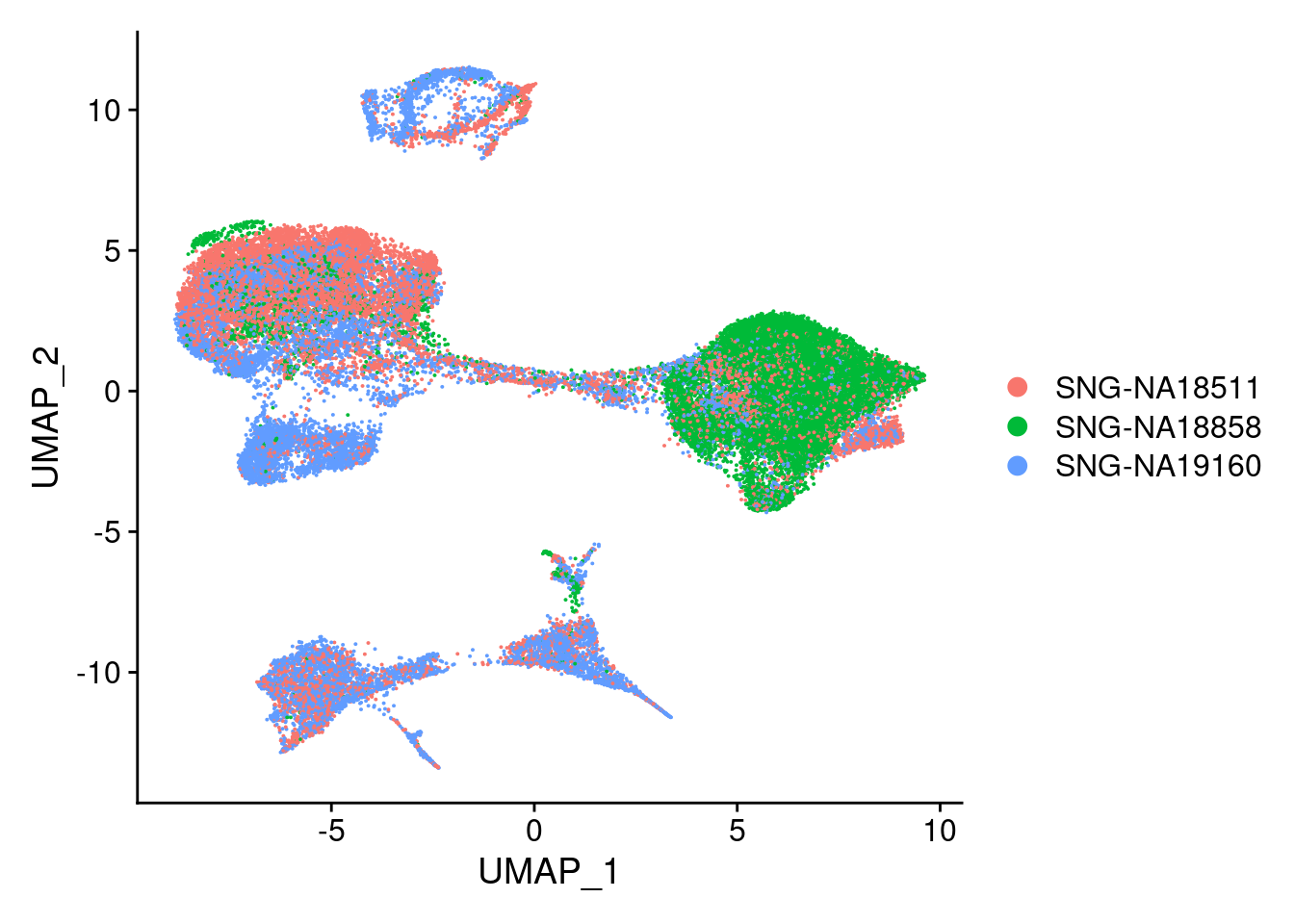
xlim <- c(min(merged@reductions$umap@cell.embeddings[,'UMAP_1']),
max(merged@reductions$umap@cell.embeddings[,'UMAP_1']))
ylim <- c(min(merged@reductions$umap@cell.embeddings[,'UMAP_2']),
max(merged@reductions$umap@cell.embeddings[,'UMAP_2']))
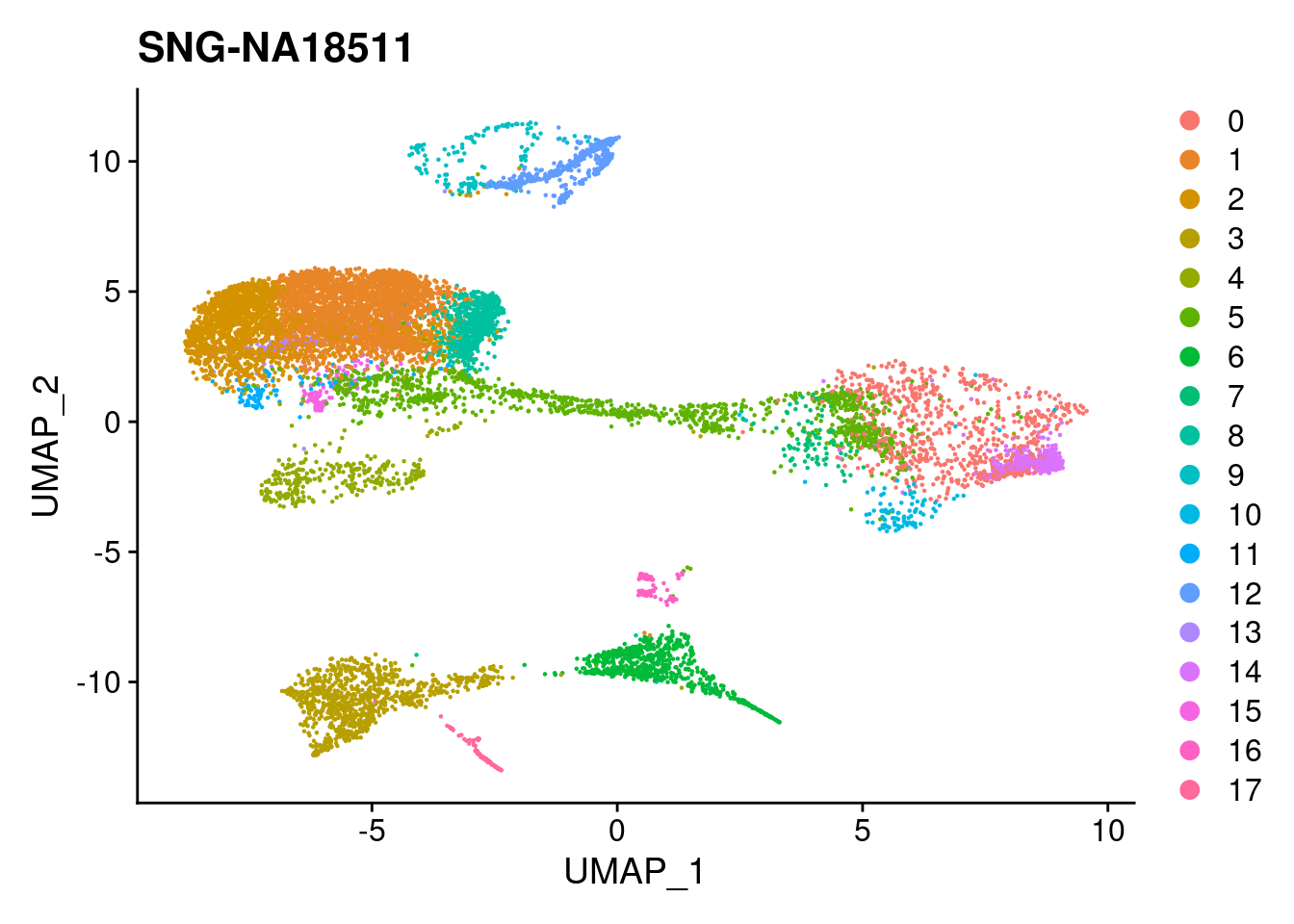
for (i in individuals)
{
print(DimPlot(merged, reduction = "umap",
cells = WhichCells(merged, expression = individual == i)) +
xlim(xlim) + ylim(ylim) + ggtitle(i))
}
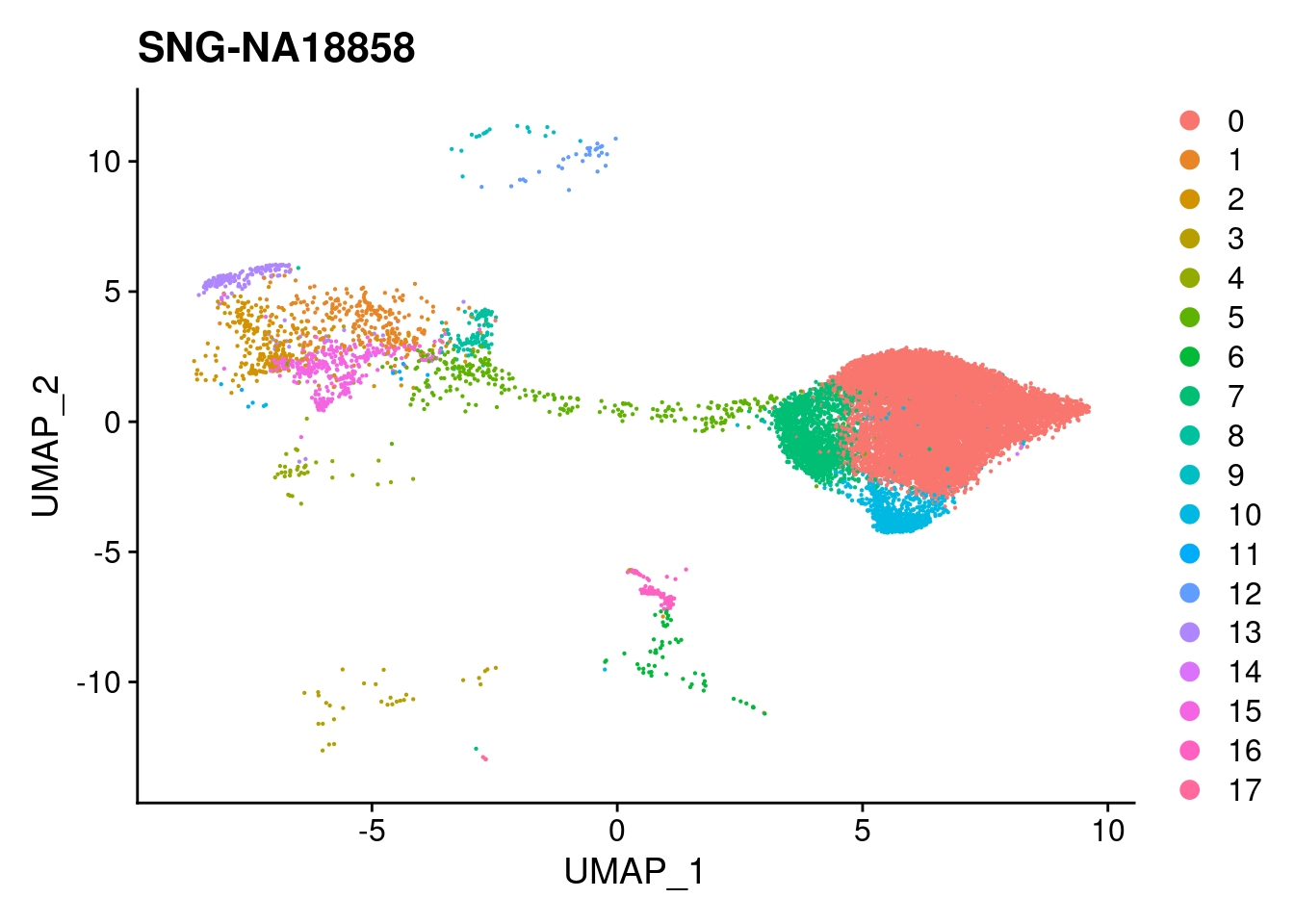
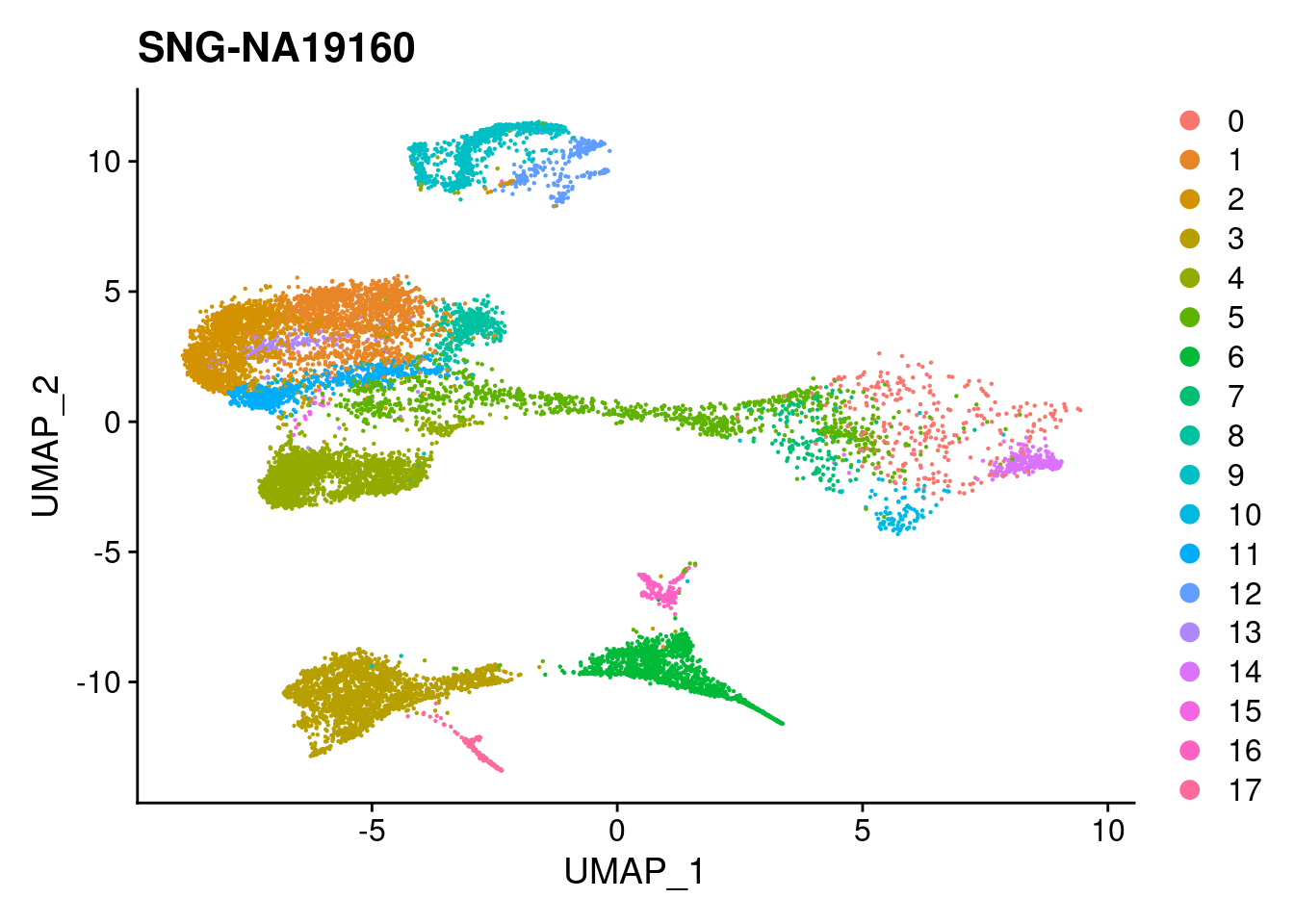
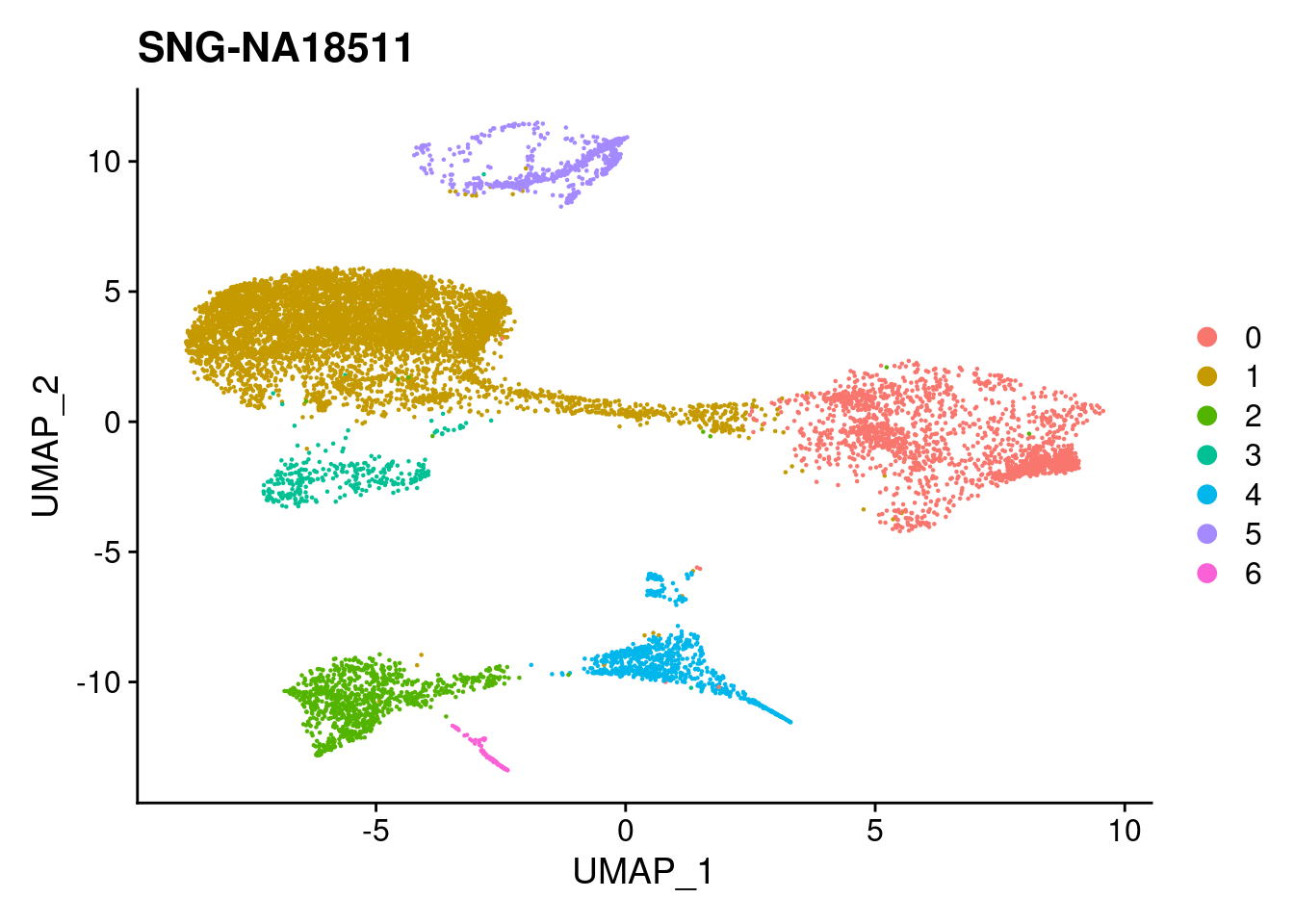
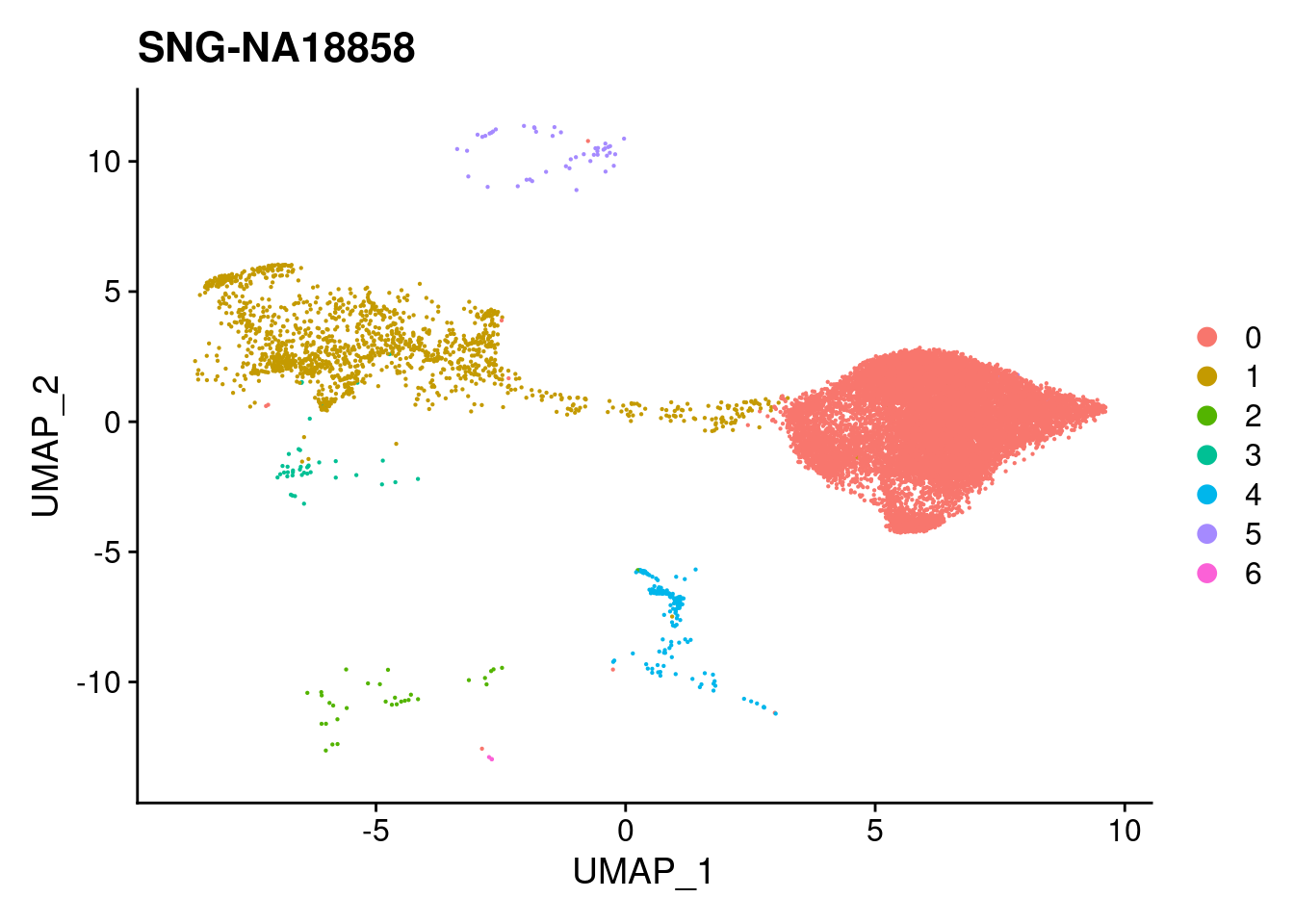
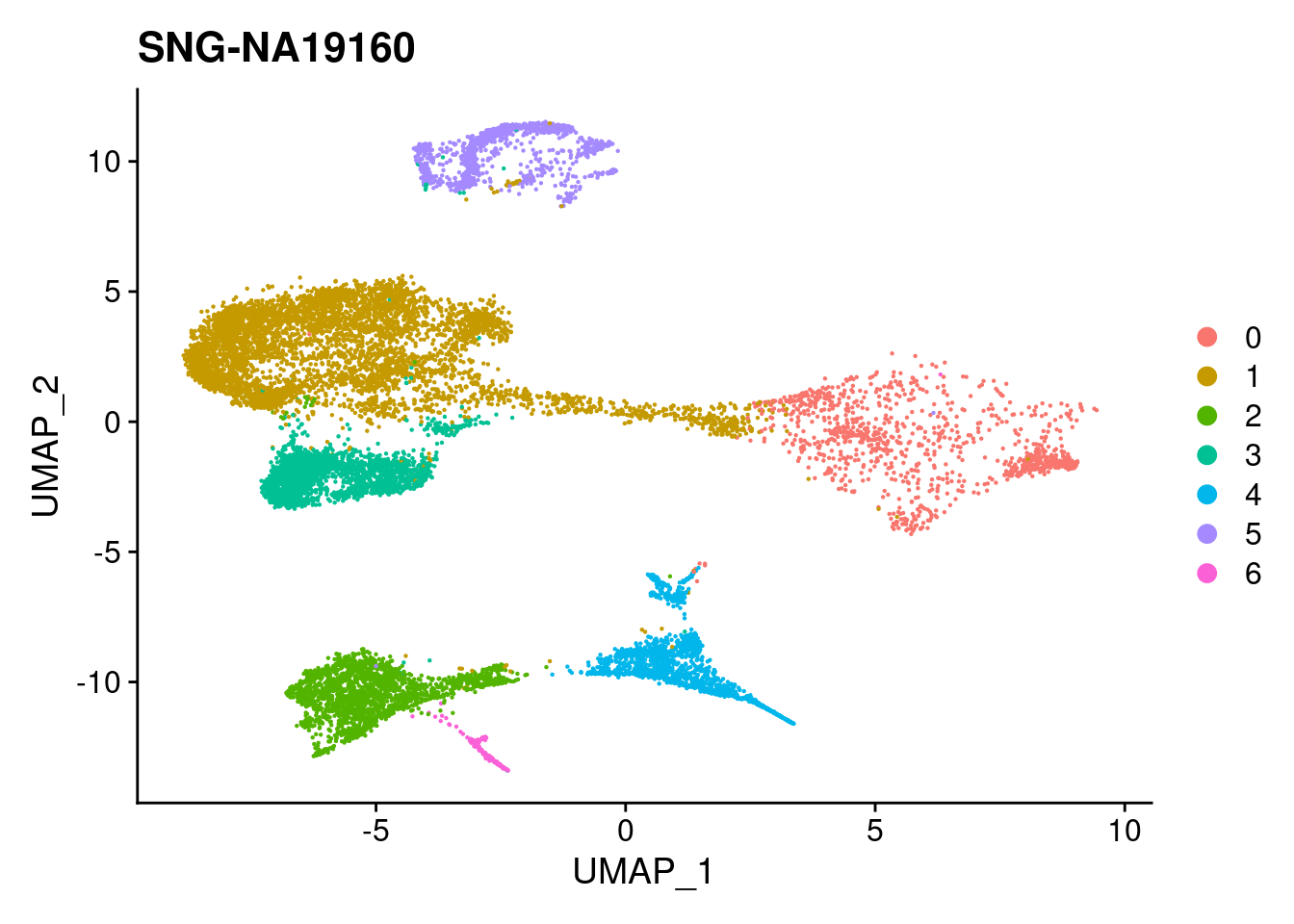
#reassign idents
Idents(merged)<- 'SCT_snn_res.0.1'DimPlot(merged, reduction = "umap")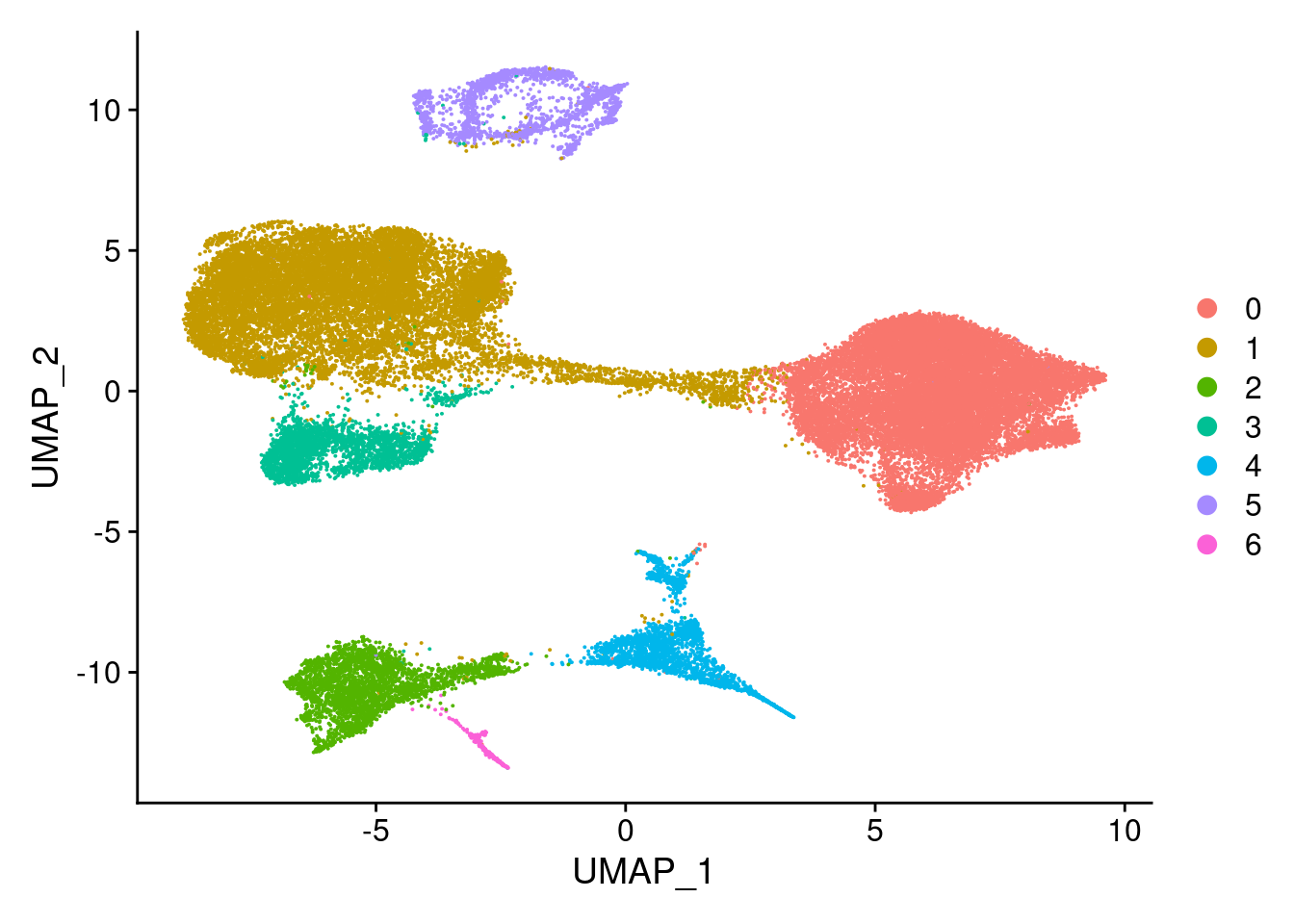
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
DimPlot(merged, reduction = "umap", group.by = "Batch")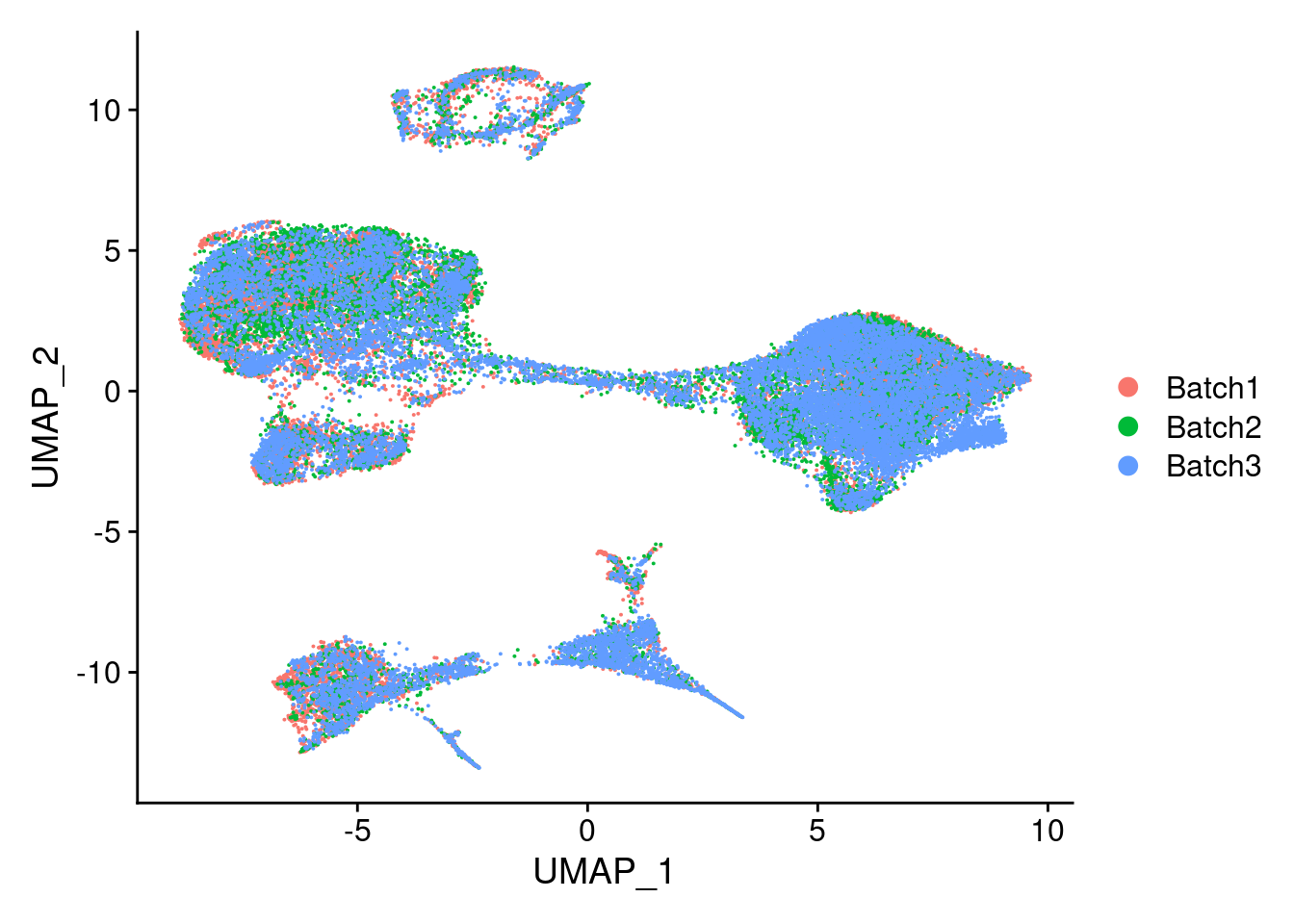
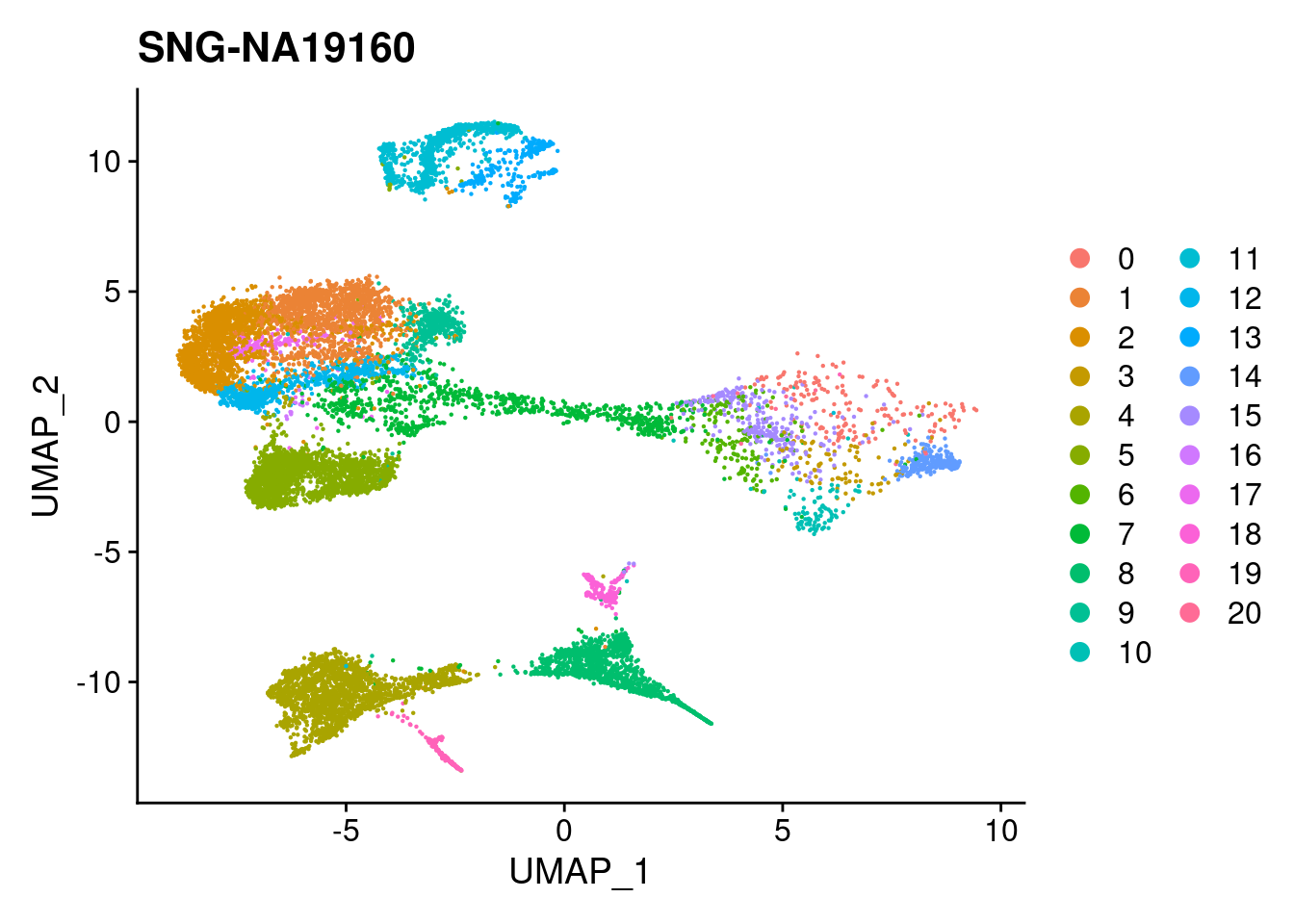
DimPlot(merged, reduction = "umap", group.by = "individual")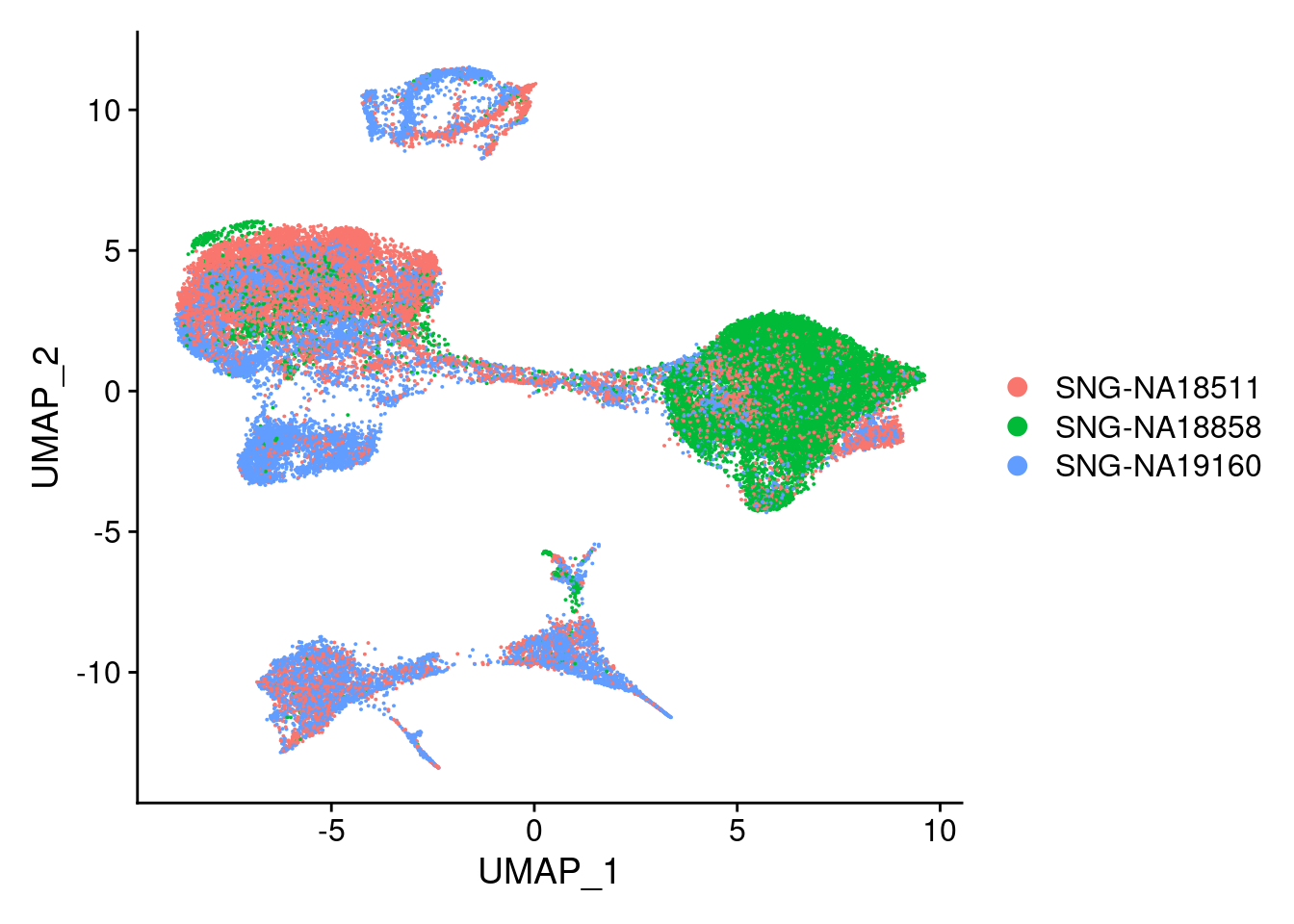
xlim <- c(min(merged@reductions$umap@cell.embeddings[,'UMAP_1']),
max(merged@reductions$umap@cell.embeddings[,'UMAP_1']))
ylim <- c(min(merged@reductions$umap@cell.embeddings[,'UMAP_2']),
max(merged@reductions$umap@cell.embeddings[,'UMAP_2']))
for (i in individuals)
{
print(DimPlot(merged, reduction = "umap",
cells = WhichCells(merged, expression = individual == i)) +
xlim(xlim) + ylim(ylim) + ggtitle(i))
}
#reassign idents
Idents(merged)<- 'SCT_snn_res.0.8'DimPlot(merged, reduction = "umap")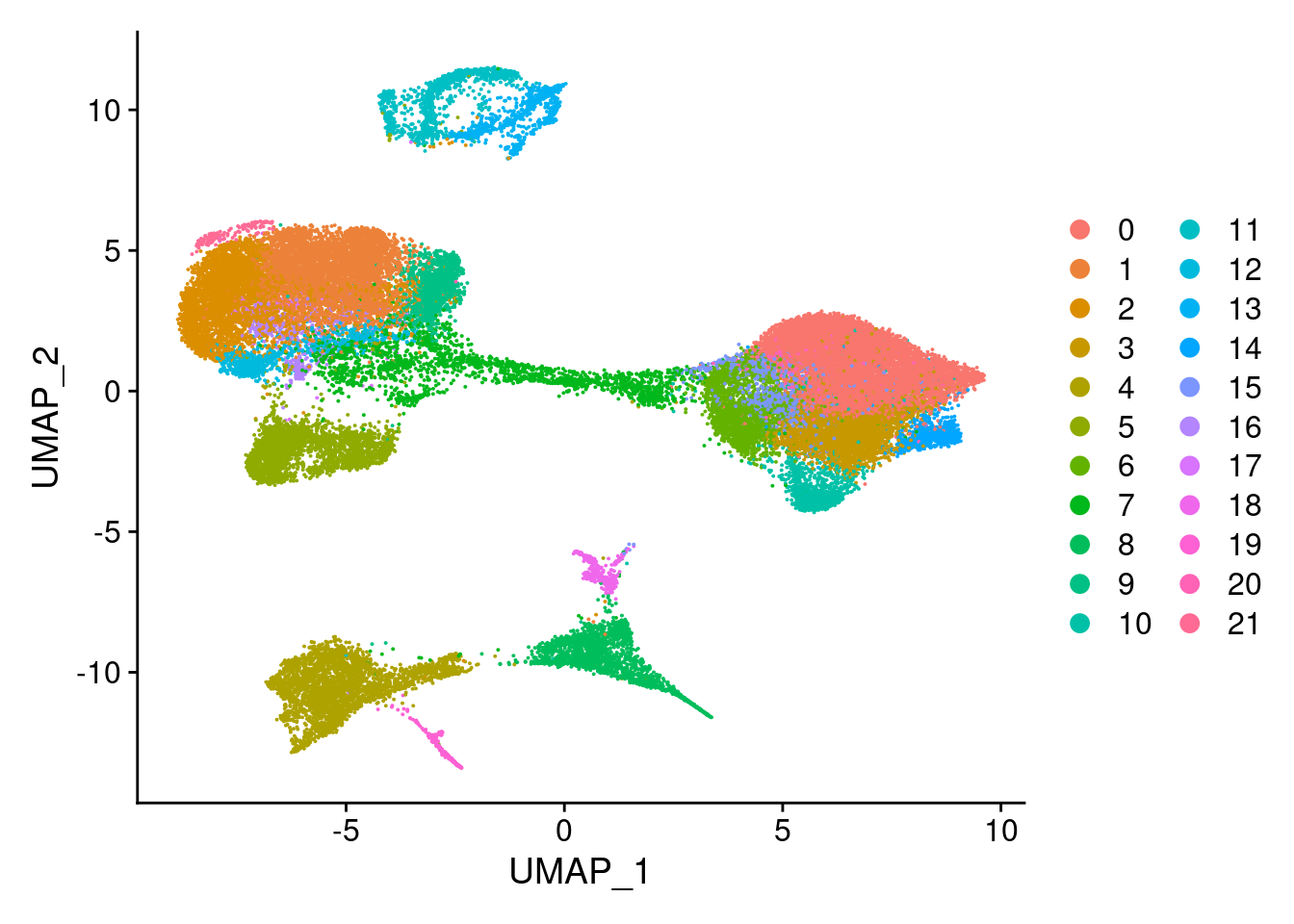
DimPlot(merged, reduction = "umap", group.by = "Batch")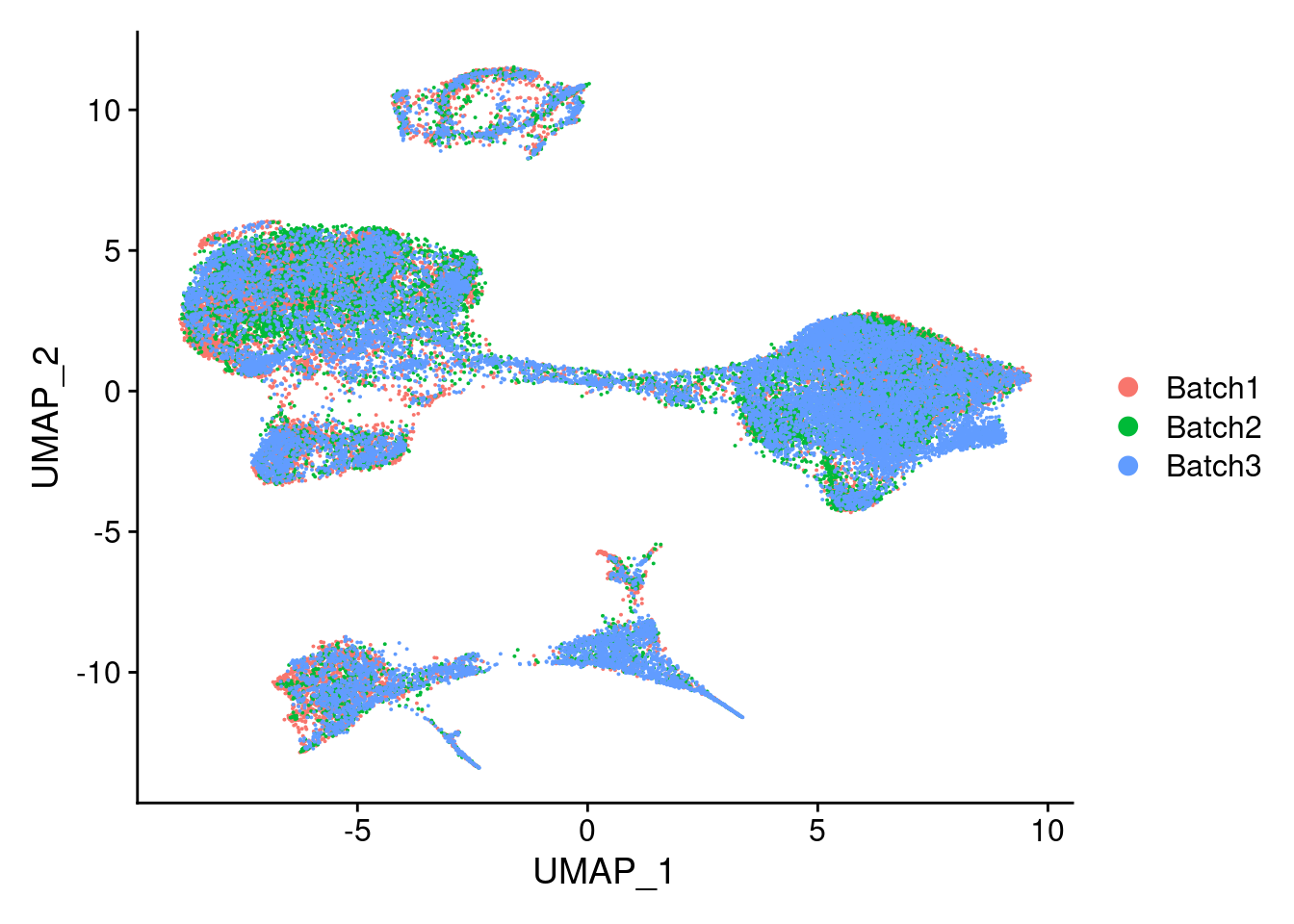
DimPlot(merged, reduction = "umap", group.by = "individual")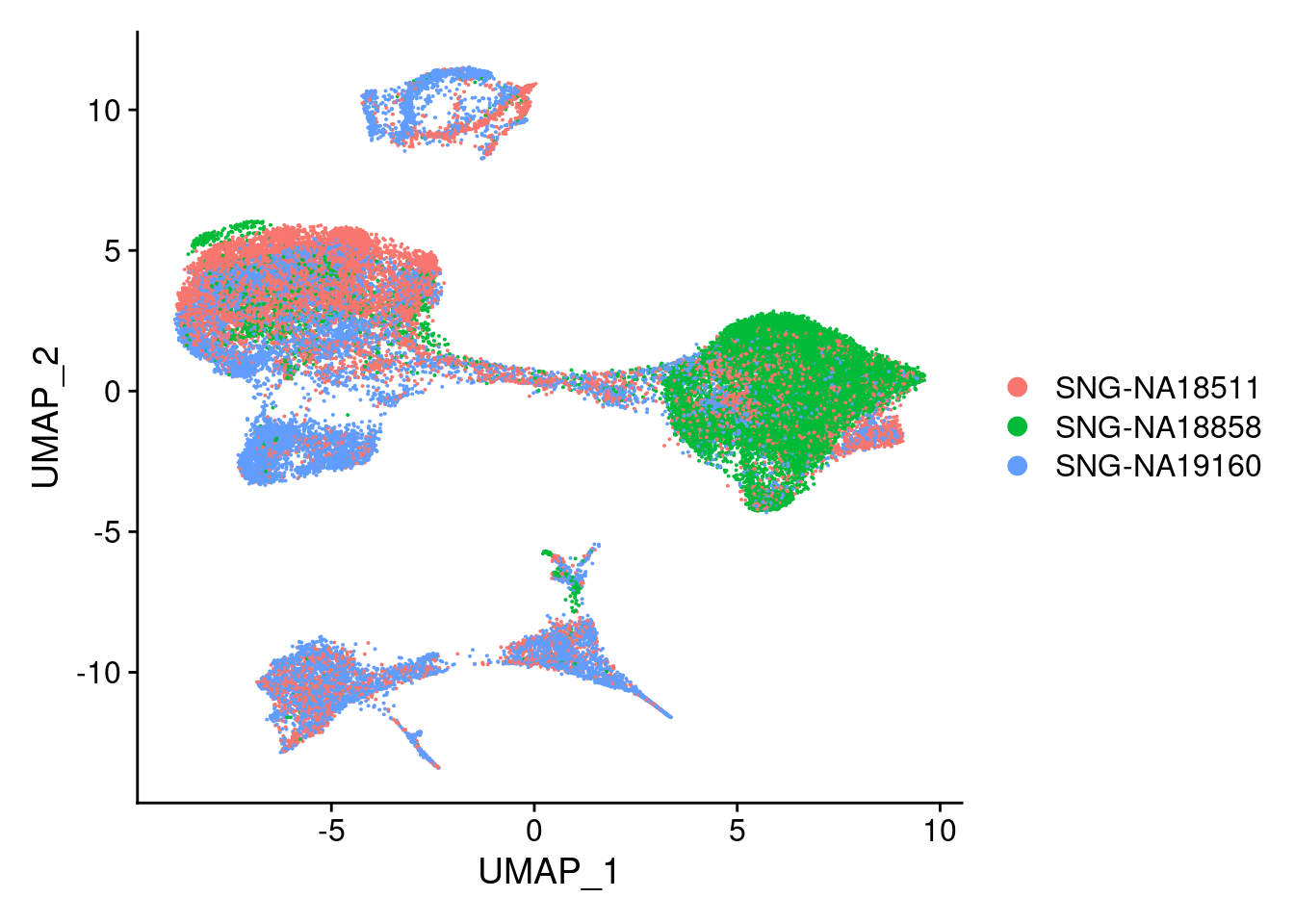
xlim <- c(min(merged@reductions$umap@cell.embeddings[,'UMAP_1']),
max(merged@reductions$umap@cell.embeddings[,'UMAP_1']))
ylim <- c(min(merged@reductions$umap@cell.embeddings[,'UMAP_2']),
max(merged@reductions$umap@cell.embeddings[,'UMAP_2']))
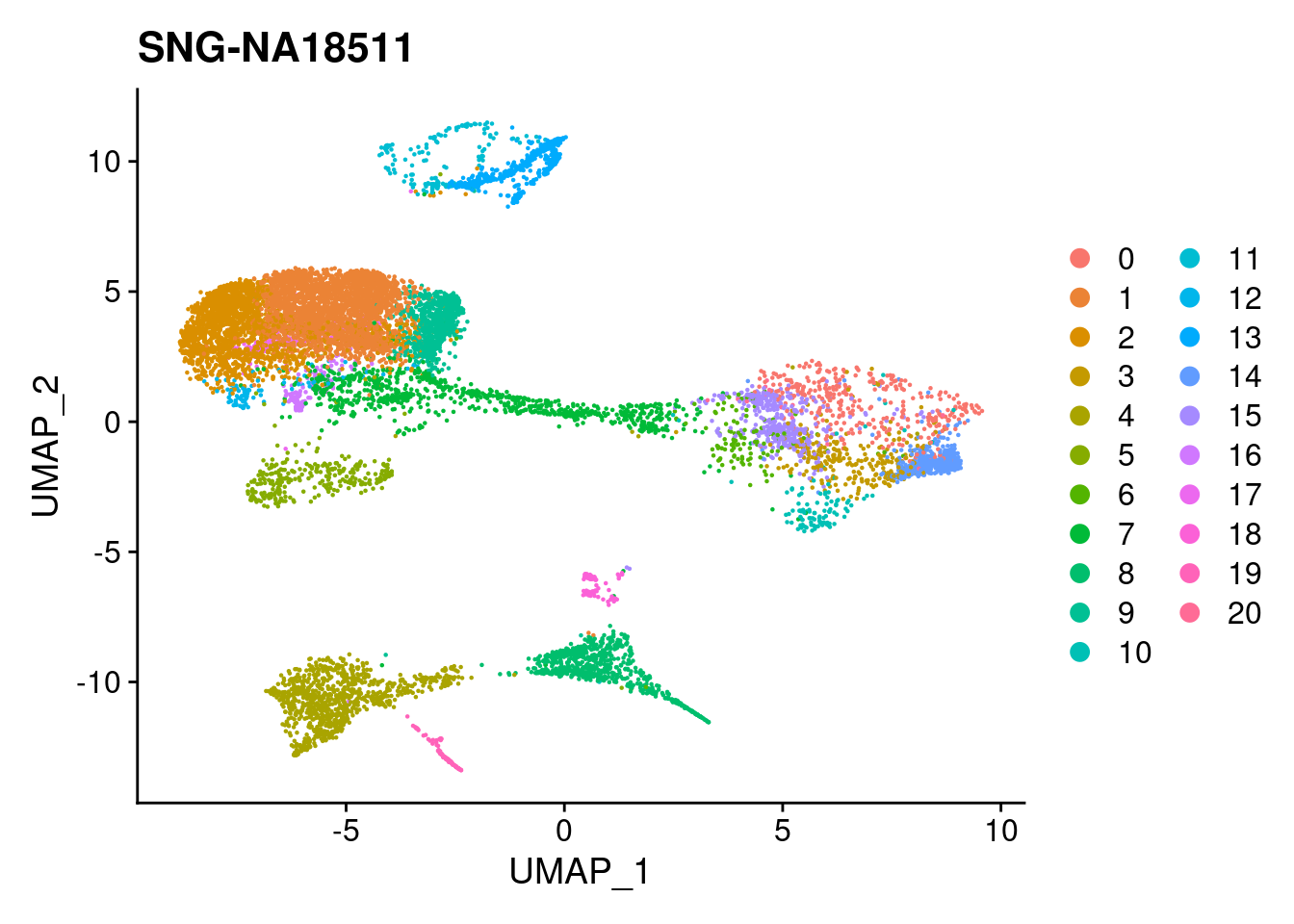
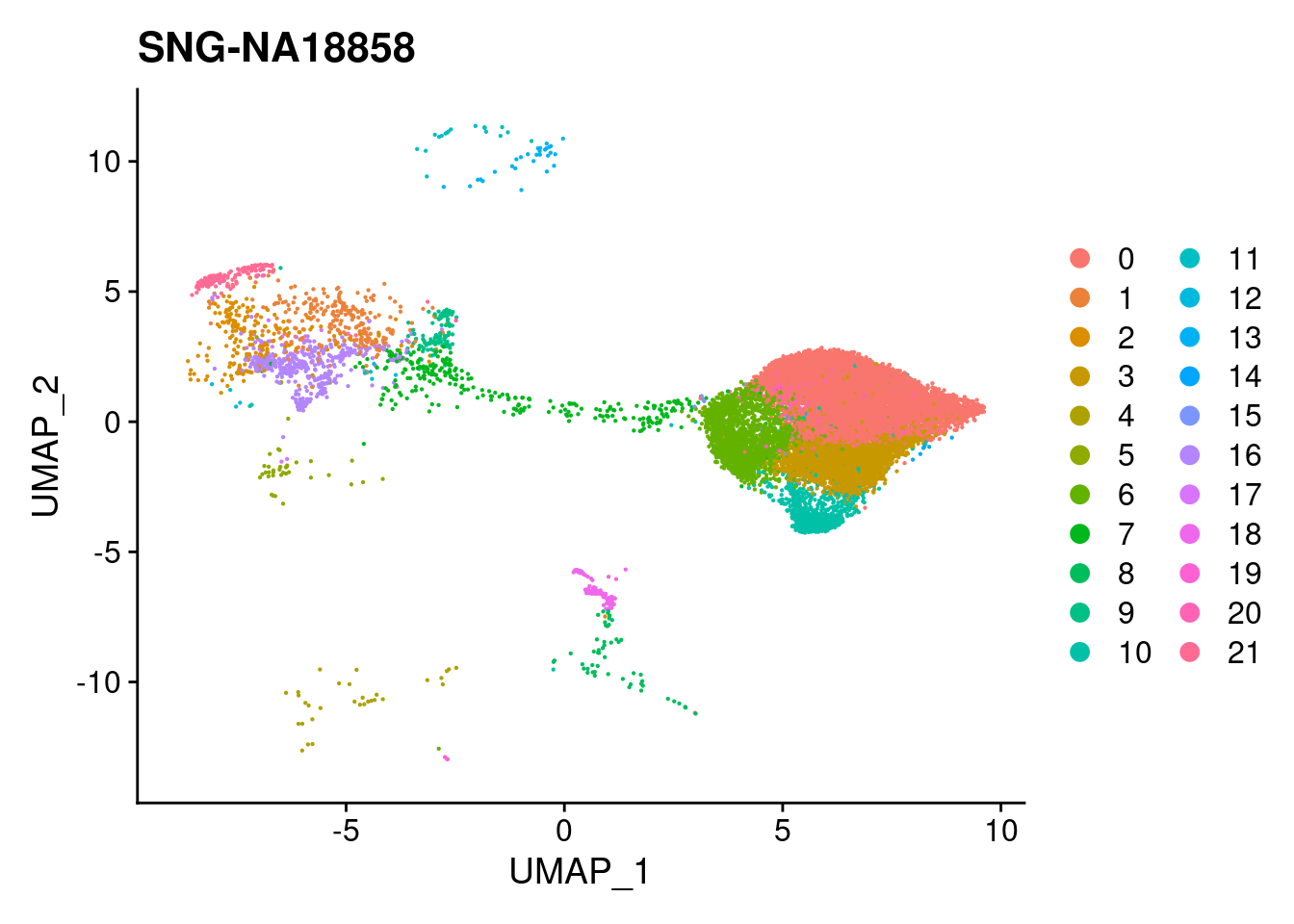
for (i in individuals)
{
print(DimPlot(merged, reduction = "umap",
cells = WhichCells(merged, expression = individual == i)) +
xlim(xlim) + ylim(ylim) + ggtitle(i))
}
VlnPlot(merged, features= "percent.mt", group.by = "SCT_snn_res.1", pt.size = 0)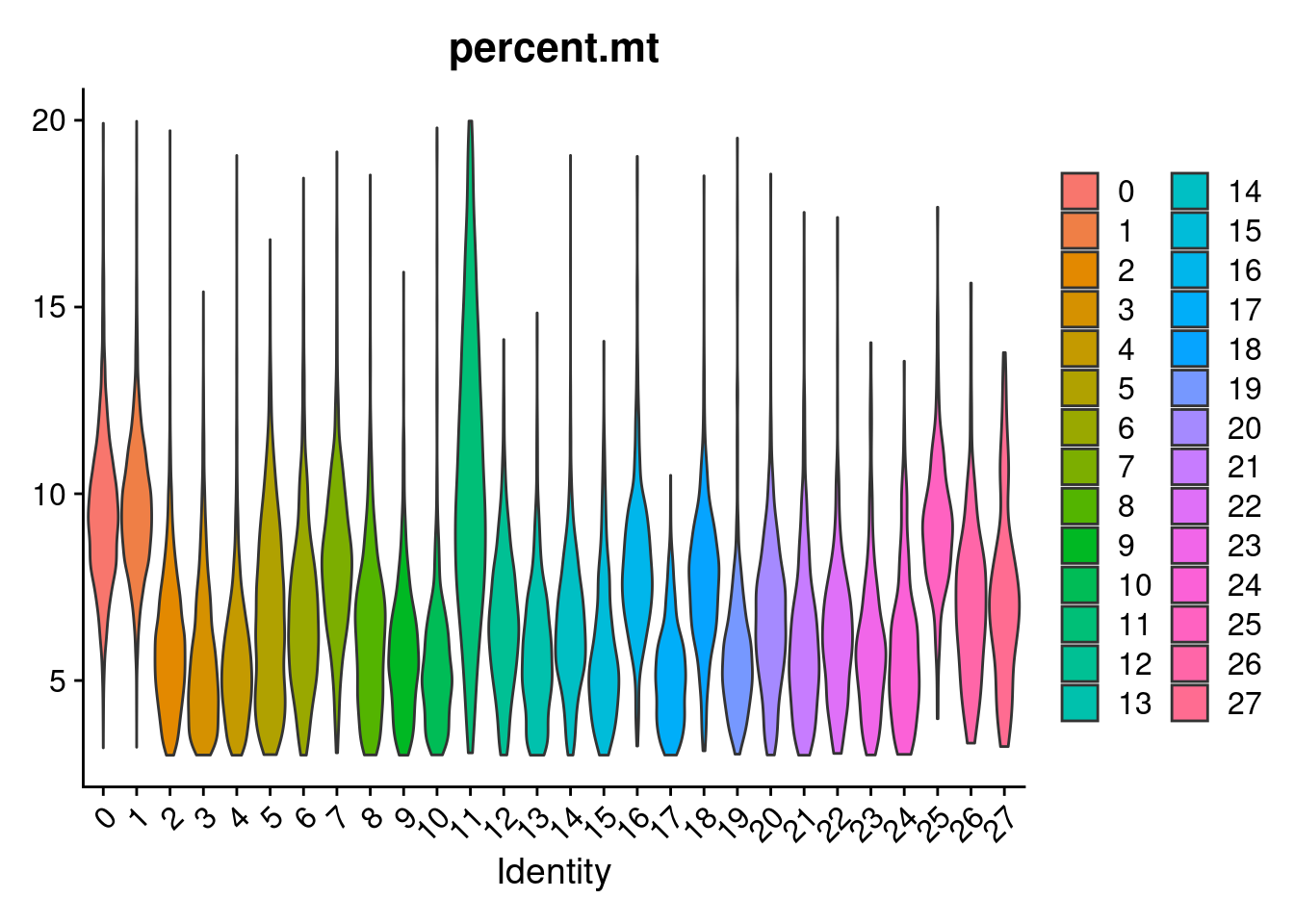
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
merged[["percent.rps"]]<- PercentageFeatureSet(merged, pattern = "^RPS")
merged[["percent.rpl"]]<- PercentageFeatureSet(merged, pattern = "^RPL")
merged[["percent.rp"]]<- merged[["percent.rps"]]+merged[["percent.rpl"]]
VlnPlot(merged, features= "percent.rp", group.by = "SCT_snn_res.1", pt.size=0)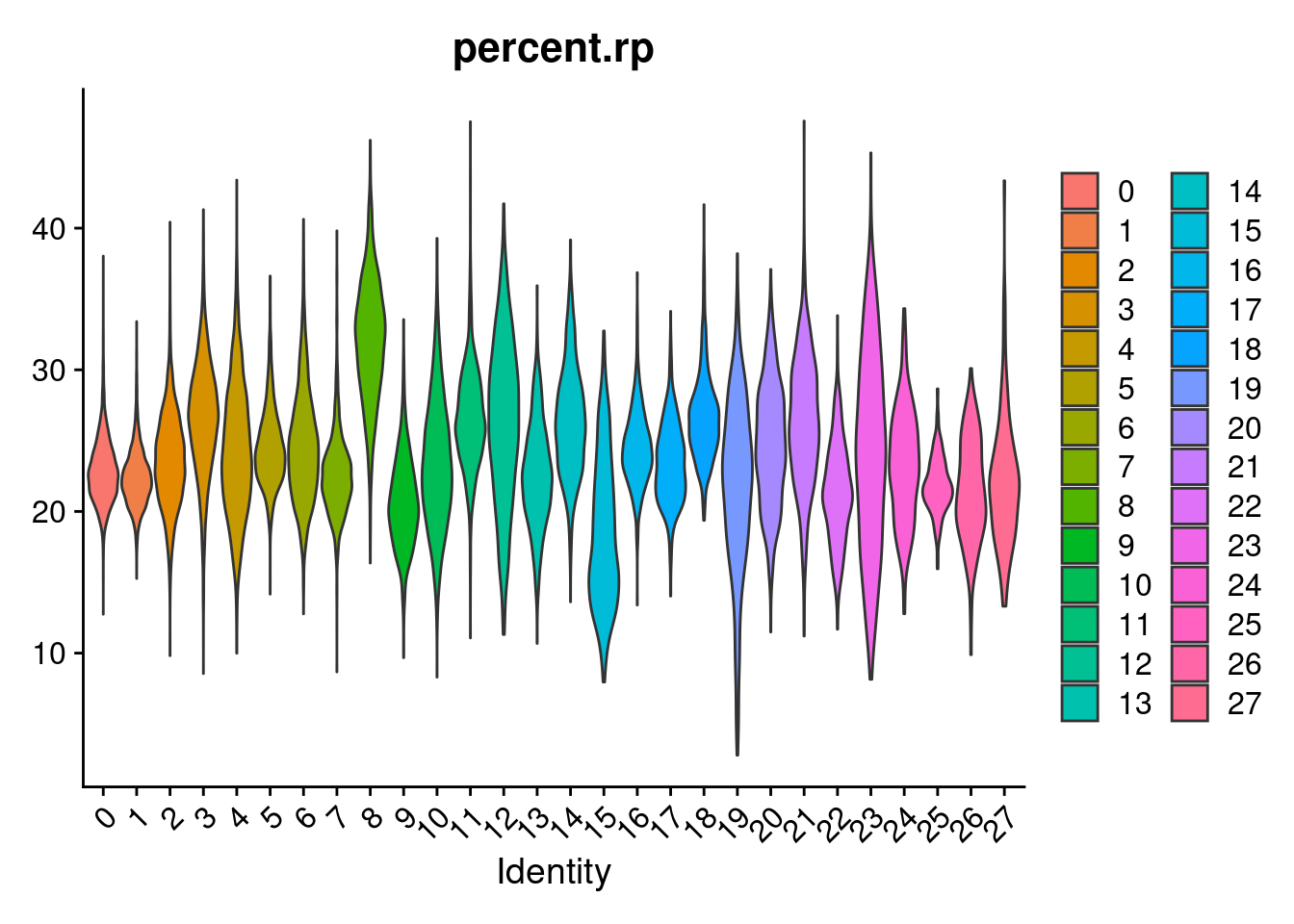
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
FeaturePlot(merged, features = "nFeature_RNA")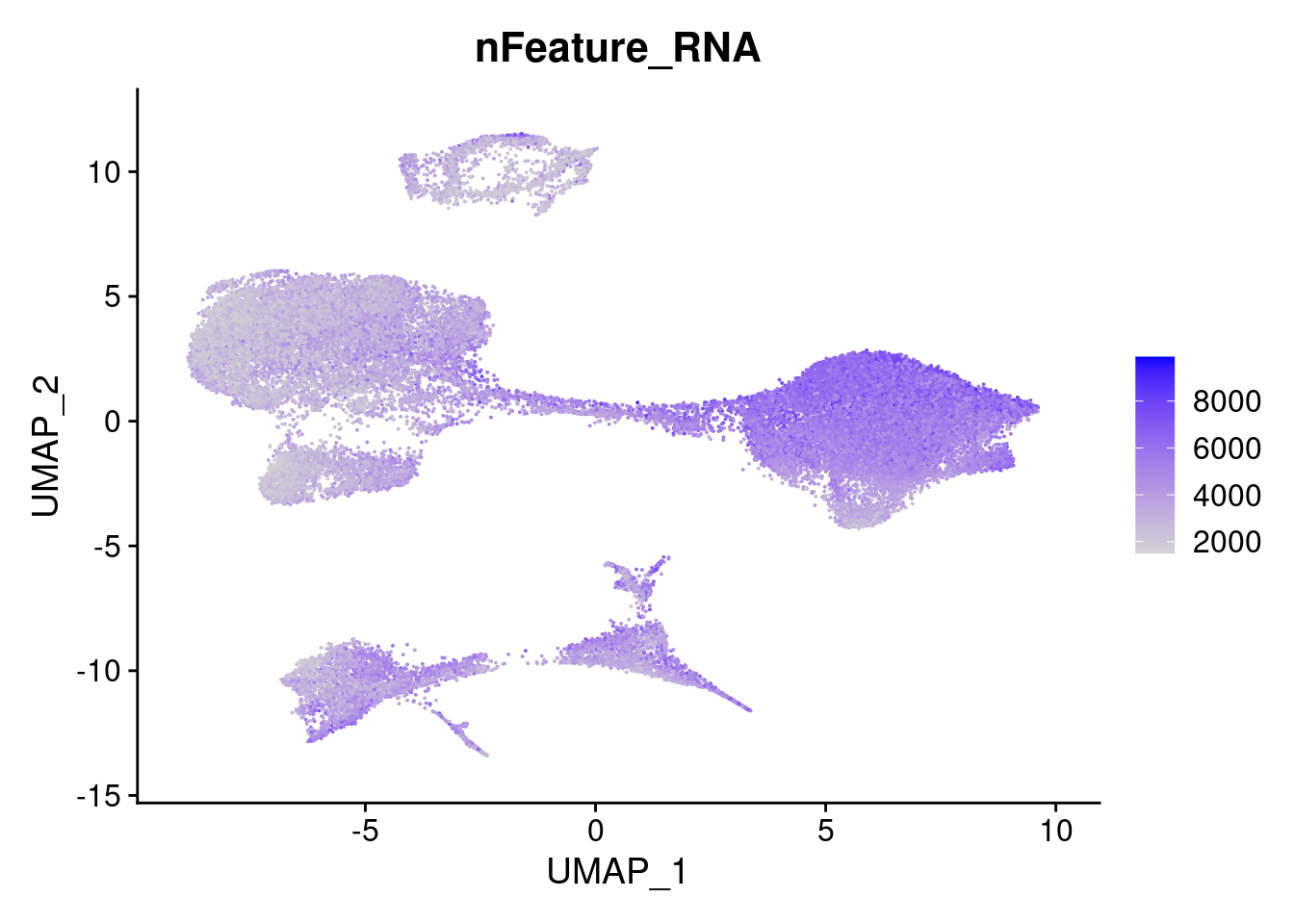
head(merged)An object of class Seurat
2 features across 42488 samples within 2 assays
Active assay: SCT (1 features, 1 variable features)
1 other assay present: RNA
3 dimensional reductions calculated: pca, harmony, umapVlnPlot(merged, features= "nFeature_RNA", group.by = "SCT_snn_res.1", pt.size=0)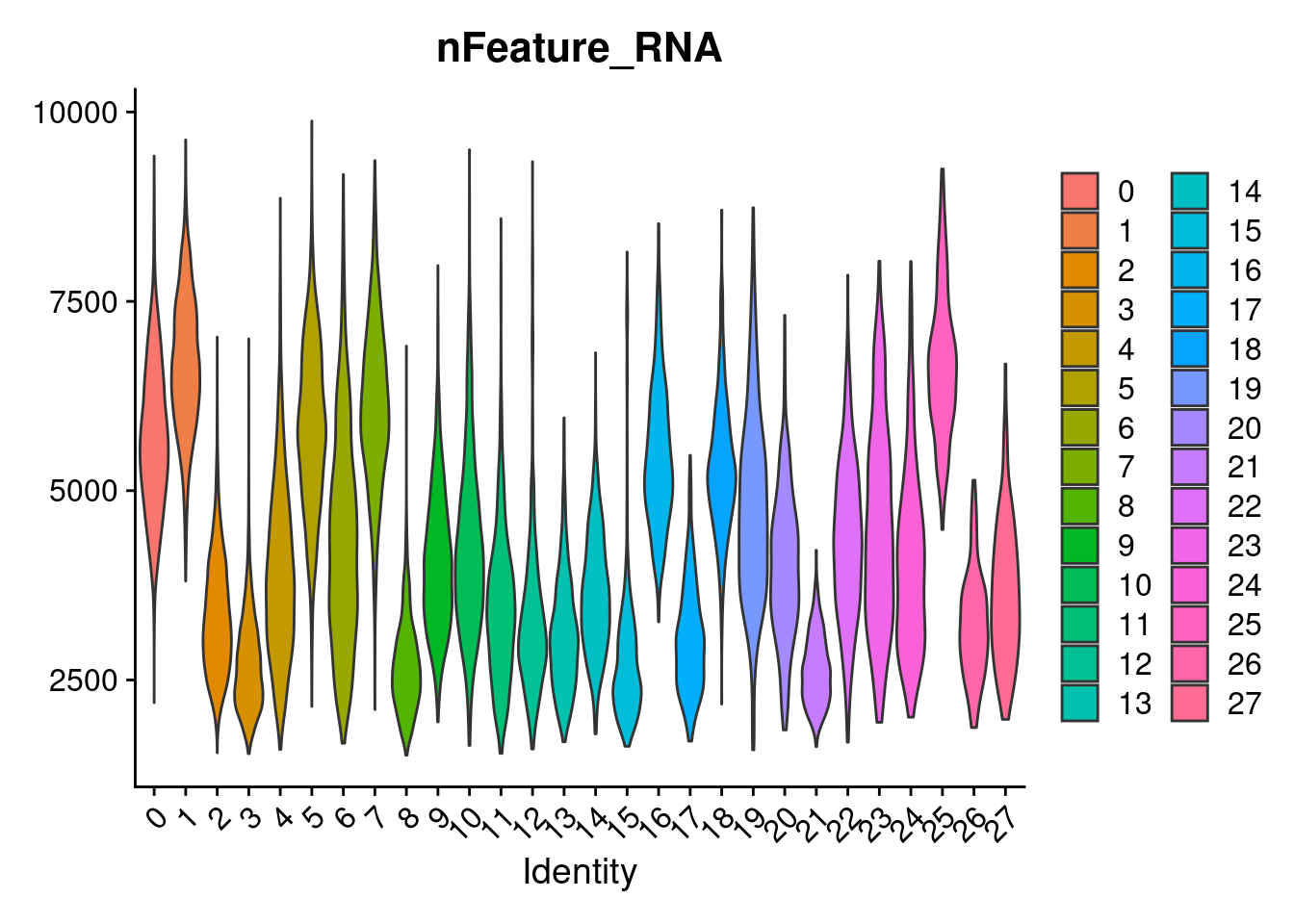
FeaturePlot(merged, features = c("POU5F1", "SOX17", "HAND1", "PAX6"), pt.size = 0.2, ncol=2, combine=T)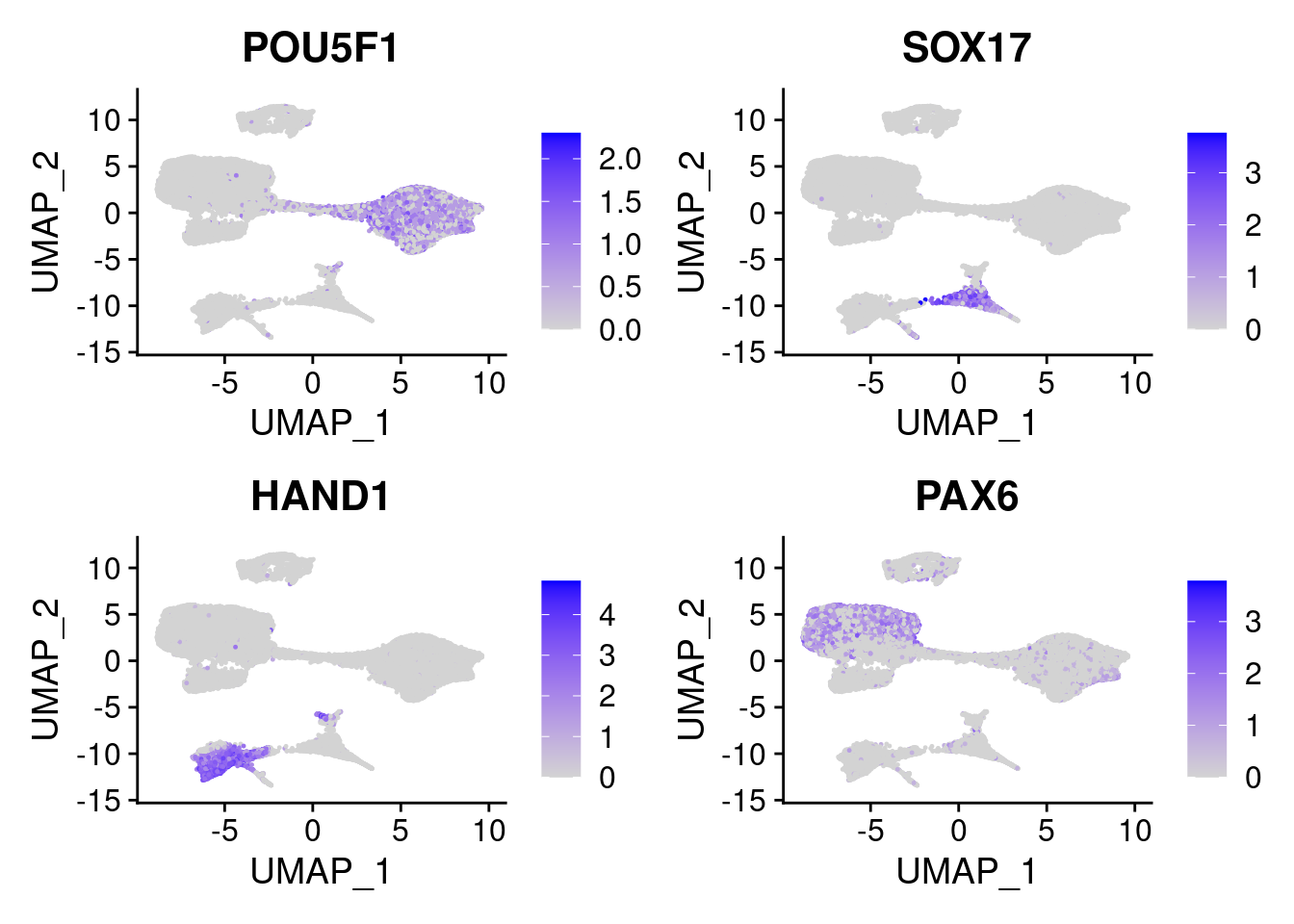
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
FeaturePlot(merged, features = c("FGB", "ECSCR", "NEUROD1", "SOX10"), pt.size = 0.2, ncol=2)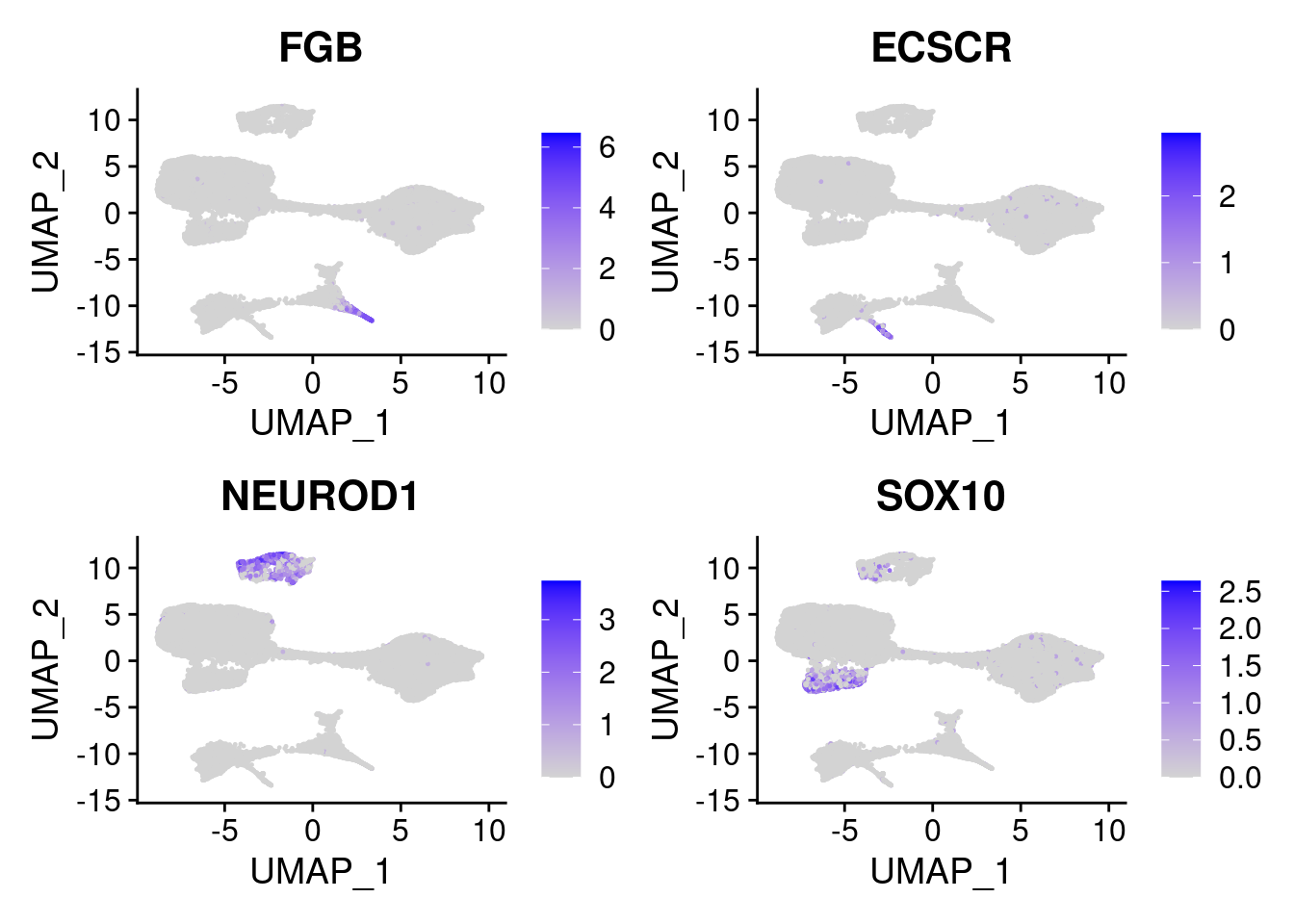
| Version | Author | Date |
|---|---|---|
| 421a225 | KLRhodes | 2020-08-10 |
sessionInfo()R version 3.6.1 (2019-07-05)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Scientific Linux 7.4 (Nitrogen)
Matrix products: default
BLAS/LAPACK: /software/openblas-0.2.19-el7-x86_64/lib/libopenblas_haswellp-r0.2.19.so
locale:
[1] C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] RColorBrewer_1.1-2 here_0.1-11 DataCombine_0.2.21 ggplot2_3.3.3
[5] harmony_1.0 Rcpp_1.0.6 Seurat_3.2.0 workflowr_1.6.2
loaded via a namespace (and not attached):
[1] Rtsne_0.15 colorspace_2.0-0 deldir_0.1-28
[4] ellipsis_0.3.1 ggridges_0.5.2 rprojroot_2.0.2
[7] fs_1.4.2 spatstat.data_1.4-3 farver_2.0.3
[10] leiden_0.3.3 listenv_0.8.0 npsurv_0.4-0
[13] ggrepel_0.9.0 RSpectra_0.16-0 codetools_0.2-16
[16] splines_3.6.1 lsei_1.2-0 knitr_1.29
[19] polyclip_1.10-0 jsonlite_1.7.2 ica_1.0-2
[22] cluster_2.1.0 png_0.1-7 uwot_0.1.10
[25] shiny_1.5.0 sctransform_0.2.1 compiler_3.6.1
[28] httr_1.4.2 Matrix_1.2-18 fastmap_1.0.1
[31] lazyeval_0.2.2 later_1.1.0.1 htmltools_0.5.0
[34] tools_3.6.1 rsvd_1.0.3 igraph_1.2.6
[37] gtable_0.3.0 glue_1.4.2 RANN_2.6.1
[40] reshape2_1.4.4 dplyr_1.0.2 rappdirs_0.3.3
[43] spatstat_1.64-1 vctrs_0.3.6 gdata_2.18.0
[46] ape_5.4-1 nlme_3.1-140 lmtest_0.9-37
[49] xfun_0.16 stringr_1.4.0 globals_0.12.5
[52] mime_0.9 miniUI_0.1.1.1 lifecycle_0.2.0
[55] irlba_2.3.3 gtools_3.8.2 goftest_1.2-2
[58] future_1.18.0 MASS_7.3-51.4 zoo_1.8-8
[61] scales_1.1.1 promises_1.1.1 spatstat.utils_1.17-0
[64] parallel_3.6.1 yaml_2.2.1 reticulate_1.20
[67] pbapply_1.4-2 gridExtra_2.3 rpart_4.1-15
[70] stringi_1.5.3 caTools_1.18.0 rlang_0.4.10
[73] pkgconfig_2.0.3 bitops_1.0-6 evaluate_0.14
[76] lattice_0.20-38 ROCR_1.0-7 purrr_0.3.4
[79] tensor_1.5 labeling_0.4.2 patchwork_1.1.1
[82] htmlwidgets_1.5.1 cowplot_1.1.1 tidyselect_1.1.0
[85] RcppAnnoy_0.0.18 plyr_1.8.6 magrittr_2.0.1
[88] R6_2.5.0 gplots_3.0.4 generics_0.1.0
[91] pillar_1.4.7 whisker_0.4 withr_2.4.2
[94] mgcv_1.8-28 fitdistrplus_1.0-14 survival_3.2-3
[97] abind_1.4-5 tibble_3.0.4 future.apply_1.6.0
[100] crayon_1.3.4 KernSmooth_2.23-15 plotly_4.9.2.1
[103] rmarkdown_2.3 grid_3.6.1 data.table_1.13.4
[106] git2r_0.26.1 digest_0.6.27 xtable_1.8-4
[109] tidyr_1.1.0 httpuv_1.5.4 munsell_0.5.0
[112] viridisLite_0.3.0
sessionInfo()R version 3.6.1 (2019-07-05)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Scientific Linux 7.4 (Nitrogen)
Matrix products: default
BLAS/LAPACK: /software/openblas-0.2.19-el7-x86_64/lib/libopenblas_haswellp-r0.2.19.so
locale:
[1] C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] RColorBrewer_1.1-2 here_0.1-11 DataCombine_0.2.21 ggplot2_3.3.3
[5] harmony_1.0 Rcpp_1.0.6 Seurat_3.2.0 workflowr_1.6.2
loaded via a namespace (and not attached):
[1] Rtsne_0.15 colorspace_2.0-0 deldir_0.1-28
[4] ellipsis_0.3.1 ggridges_0.5.2 rprojroot_2.0.2
[7] fs_1.4.2 spatstat.data_1.4-3 farver_2.0.3
[10] leiden_0.3.3 listenv_0.8.0 npsurv_0.4-0
[13] ggrepel_0.9.0 RSpectra_0.16-0 codetools_0.2-16
[16] splines_3.6.1 lsei_1.2-0 knitr_1.29
[19] polyclip_1.10-0 jsonlite_1.7.2 ica_1.0-2
[22] cluster_2.1.0 png_0.1-7 uwot_0.1.10
[25] shiny_1.5.0 sctransform_0.2.1 compiler_3.6.1
[28] httr_1.4.2 Matrix_1.2-18 fastmap_1.0.1
[31] lazyeval_0.2.2 later_1.1.0.1 htmltools_0.5.0
[34] tools_3.6.1 rsvd_1.0.3 igraph_1.2.6
[37] gtable_0.3.0 glue_1.4.2 RANN_2.6.1
[40] reshape2_1.4.4 dplyr_1.0.2 rappdirs_0.3.3
[43] spatstat_1.64-1 vctrs_0.3.6 gdata_2.18.0
[46] ape_5.4-1 nlme_3.1-140 lmtest_0.9-37
[49] xfun_0.16 stringr_1.4.0 globals_0.12.5
[52] mime_0.9 miniUI_0.1.1.1 lifecycle_0.2.0
[55] irlba_2.3.3 gtools_3.8.2 goftest_1.2-2
[58] future_1.18.0 MASS_7.3-51.4 zoo_1.8-8
[61] scales_1.1.1 promises_1.1.1 spatstat.utils_1.17-0
[64] parallel_3.6.1 yaml_2.2.1 reticulate_1.20
[67] pbapply_1.4-2 gridExtra_2.3 rpart_4.1-15
[70] stringi_1.5.3 caTools_1.18.0 rlang_0.4.10
[73] pkgconfig_2.0.3 bitops_1.0-6 evaluate_0.14
[76] lattice_0.20-38 ROCR_1.0-7 purrr_0.3.4
[79] tensor_1.5 labeling_0.4.2 patchwork_1.1.1
[82] htmlwidgets_1.5.1 cowplot_1.1.1 tidyselect_1.1.0
[85] RcppAnnoy_0.0.18 plyr_1.8.6 magrittr_2.0.1
[88] R6_2.5.0 gplots_3.0.4 generics_0.1.0
[91] pillar_1.4.7 whisker_0.4 withr_2.4.2
[94] mgcv_1.8-28 fitdistrplus_1.0-14 survival_3.2-3
[97] abind_1.4-5 tibble_3.0.4 future.apply_1.6.0
[100] crayon_1.3.4 KernSmooth_2.23-15 plotly_4.9.2.1
[103] rmarkdown_2.3 grid_3.6.1 data.table_1.13.4
[106] git2r_0.26.1 digest_0.6.27 xtable_1.8-4
[109] tidyr_1.1.0 httpuv_1.5.4 munsell_0.5.0
[112] viridisLite_0.3.0