Limma Res 0.8
Katie Rhodes
5/27/2021
Last updated: 2021-12-13
Checks: 6 1
Knit directory: Embryoid_Body_Pilot_Workflowr/analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it's best to always run the code in an empty environment.
The command set.seed(20200804) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Using absolute paths to the files within your workflowr project makes it difficult for you and others to run your code on a different machine. Change the absolute path(s) below to the suggested relative path(s) to make your code more reproducible.
| absolute | relative |
|---|---|
| /project2/gilad/katie/Pilot_HumanEBs/Embryoid_Body_Pilot_Workflowr/output/NEW_Pseudobulk_Limma_res0.8_OnevAllTopTables.csv | ../output/NEW_Pseudobulk_Limma_res0.8_OnevAllTopTables.csv |
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version e6309e4. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: analysis/.Rhistory
Ignored: output/.Rhistory
Untracked files:
Untracked: GSE122380_raw_counts.txt.gz
Untracked: UTF1_plots.Rmd
Untracked: analysis/DoubletFinderTest.Rmd
Untracked: analysis/IntegrateReference_SCTregressCaoPlusScHCL_5newAndOriginal3.Rmd
Untracked: analysis/OLD/
Untracked: analysis/child/
Untracked: build_refint_5new.R
Untracked: build_refint_scale.R
Untracked: build_refint_sct.R
Untracked: build_stuff.R
Untracked: build_varpart_sc.R
Untracked: code/.ipynb_checkpoints/
Untracked: code/CellRangerPreprocess.Rmd
Untracked: code/GEO_processed_data.Rmd
Untracked: code/GEO_processed_data_additionalLines.Rmd
Untracked: code/PowerAnalysis_NoiseRatio.ipynb
Untracked: code/Rplots.pdf
Untracked: code/Untitled.ipynb
Untracked: code/Untitled1.ipynb
Untracked: data/HCL_Fig1_adata.h5ad
Untracked: data/HCL_Fig1_adata.h5seurat
Untracked: data/dge/
Untracked: data/dge_raw_data.tar.gz
Untracked: data/ref.expr.rda
Untracked: figure/
Untracked: output/5NEWLINES.BroadCellTypeCatAssignment.basedonclustersres0.15.csv
Untracked: output/5NEWLINES.Frequency.MostCommonAnnotation.FiveNearestRefCells.csv
Untracked: output/5NEWLINES.MostCommonAnnotation.FiveNearestRefCells.csv
Untracked: output/5NEWLINES.NearestReferenceCell.Cao.hESC.EuclideanDistanceinHarmonySpace.csv
Untracked: output/5NEWLINES.NearestReferenceCell.Cao.hESC.FrequencyofEachAnnotation.csv
Untracked: output/5newlines.merge.all.SCTwRegressOrigIdent.Harmony.rds
Untracked: output/CR_sampleQCrds/
Untracked: output/CaoEtAl.Obj.CellsOfAllClusters.ProteinCodingGenes.rds
Untracked: output/CaoEtAl.Obj.rds
Untracked: output/ClusterInfo_res0.1.csv
Untracked: output/DGELists/
Untracked: output/DoubletFinderTestSweepResults.RData
Untracked: output/DoubletFinderTestWITHknowndoublets.RData
Untracked: output/DownSampleVarPart.rds
Untracked: output/FiveNewLinesBarcodes.csv
Untracked: output/Frequency.MostCommonAnnotation.FiveNearestRefCells.csv
Untracked: output/GEOsubmissionProcessedFiles/
Untracked: output/GEOsubmission_additionalLines/
Untracked: output/GeneLists_by_minPCT/
Untracked: output/MostCommonAnnotation.FiveNearestRefCells.csv
Untracked: output/NEW_Pseudobulk_Limma_res0.1_OnevAllTopTables.csv
Untracked: output/NEW_Pseudobulk_Limma_res0.5_OnevAllTopTables.csv
Untracked: output/NEW_Pseudobulk_Limma_res0.8_OnevAllTopTables.csv
Untracked: output/NEW_Pseudobulk_Limma_res1_OnevAllTopTables.csv
Untracked: output/NearestReferenceCell.Cao.hESC.EuclideanDistanceinHarmonySpace.csv
Untracked: output/NearestReferenceCell.Cao.hESC.FrequencyofEachAnnotation.csv
Untracked: output/NearestReferenceCell.SCTregressRNAassay.Cao.hESC.EuclideanDistanceinHarmonySpace.csv
Untracked: output/NearestReferenceCell.SCTregressRNAassay.Cao.hESC.FrequencyofEachAnnotation.csv
Untracked: output/NewAndOriginal.merge.all.SCTwRegressOrigIdent.Harmony.rds
Untracked: output/Pseudobulk_Limma_res0.1_OnevAllTopTables.csv
Untracked: output/Pseudobulk_Limma_res0.1_OnevAll_top10Upregby_adjP.csv
Untracked: output/Pseudobulk_Limma_res0.1_OnevAll_top10Upregby_logFC.csv
Untracked: output/Pseudobulk_Limma_res0.5_OnevAllTopTables.csv
Untracked: output/Pseudobulk_Limma_res0.8_OnevAllTopTables.csv
Untracked: output/Pseudobulk_Limma_res1_OnevAllTopTables.csv
Untracked: output/Pseudobulk_VarPart.ByCluster.Res0.1.rds
Untracked: output/ResidualVariances_fromDownSampAnalysis.csv
Untracked: output/SingleCell_VariancePartition_RNA_Res0.1_minPCT0.2.rds
Untracked: output/SingleCell_VariancePartition_Res0.1_minPCT0.2.rds
Untracked: output/SingleCell_VariancePartition_SCT_Res0.1_minPCT0.2.rds
Untracked: output/TopicModelling_k10_top10drivergenes.byBeta.csv
Untracked: output/TopicModelling_k6_top10drivergenes.byBeta.csv
Untracked: output/TopicModelling_k6_top15drivergenes.byZ.csv
Untracked: output/TranferredAnnotations_ReferenceInt_JustEarlyEcto.csv
Untracked: output/TranferredAnnotations_ReferenceInt_JustEndoderm.csv
Untracked: output/TranferredAnnotations_ReferenceInt_JustMeso.csv
Untracked: output/TranferredAnnotations_ReferenceInt_JustNeuralCrest.csv
Untracked: output/TranferredAnnotations_ReferenceInt_JustNeuron.csv
Untracked: output/TranferredAnnotations_ReferenceInt_JustPluripotent.csv
Untracked: output/VarPart.ByCluster.Res0.1.rds
Untracked: output/azimuth/
Untracked: output/downsamp_10800cells_10subreps_medianexplainedbyresiduals_varpart_PsB.rds
Untracked: output/downsamp_16200cells_10subreps_medianexplainedbyresiduals_varpart_PsB.rds
Untracked: output/downsamp_21600cells_10subreps_medianexplainedbyresiduals_varpart_PsB.rds
Untracked: output/downsamp_2700cells_10subreps_medianexplainedbyresiduals_varpart_PsB.rds
Untracked: output/downsamp_2700cells_10subreps_medianexplainedbyresiduals_varpart_scres.rds
Untracked: output/downsamp_5400cells_10subreps_medianexplainedbyresiduals_varpart_PsB.rds
Untracked: output/downsamp_7200cells_10subreps_medianexplainedbyresiduals_varpart_PsB.rds
Untracked: output/fasttopics/
Untracked: output/figs/
Untracked: output/merge.Cao.SCTwRegressOrigIdent.rds
Untracked: output/merge.all.SCTwRegressOrigIdent.Harmony.rds
Untracked: output/merged.SCT.counts.matrix.rds
Untracked: output/merged.raw.counts.matrix.rds
Untracked: output/mergedObjects/
Untracked: output/pdfs/
Untracked: output/sampleQCrds/
Untracked: output/splitgpm_gsea_results/
Untracked: publish_stuff1.R
Untracked: publish_stuff2.R
Untracked: slurm-14911987.out
Unstaged changes:
Deleted: analysis/IntegrateAnalysis.afterFilter.HarmonyBatch.Rmd
Deleted: analysis/IntegrateAnalysis.afterFilter.HarmonyBatchSampleIDindividual.Rmd
Deleted: analysis/IntegrateAnalysis.afterFilter.NOHARMONYjustmerge.Rmd
Deleted: analysis/IntegrateAnalysis.afterFilter.SCTregressBatchIndividual.Rmd
Deleted: analysis/IntegrateAnalysis.afterFilter.SCTregressBatchIndividualHarmonyBatchindividual.Rmd
Modified: analysis/Pseudobulk_Limma_Harmony.BatchIndividual_ClusterRes0.1_minPCT0.2.Rmd
Modified: analysis/Pseudobulk_Limma_Harmony.BatchIndividual_ClusterRes1_minPCT0.2.Rmd
Deleted: analysis/RunscHCL_HarmonyBatchInd.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/Pseudobulk_Limma_Harmony.BatchIndividual_ClusterRes0.8_minPCT0.2.Rmd) and HTML (docs/Pseudobulk_Limma_Harmony.BatchIndividual_ClusterRes0.8_minPCT0.2.html) files. If you've configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | e6309e4 | KLRhodes | 2021-12-13 | Fix supp files |
| Rmd | fe7fae8 | KLRhodes | 2021-07-05 | wflow_git_commit(c("analysis/Pseudobulk_HierarchicalClustering_Harmony.Batchindividual_ClusterRes*", |
| html | 475b623 | KLRhodes | 2021-07-05 | Build site. |
library(Seurat)
library(Matrix)
library(dplyr)
library(edgeR)
library(limma)
library(tibble)choose parameters (integration type, clustering res, min pct threshold)
f<- 'Harmony.Batchindividual'
pct<-0.2
res<- 'SCT_snn_res.0.8'path<- here::here("output/DGELists/")
dge<- readRDS(paste0(path,"Pseudobulk_dge_",f, "_", res,"_minPCT",pct,".rds"))genes.ribo <- grep('^RP',rownames(dge),value=T)
genes.no.ribo <- rownames(dge)[which(!(rownames(dge) %in% genes.ribo))]
dge$counts <- dge$counts[which(rownames(dge$counts) %in% genes.no.ribo),] #remove ribosomal genesdge<- calcNormFactors(dge, method="TMM")
summary(dge$samples$norm.factors) Min. 1st Qu. Median Mean 3rd Qu. Max.
0.7666 0.9011 0.9717 1.0158 1.0588 2.0554 design<- model.matrix(~0+ dge$samples$cluster + dge$samples$batch + dge$samples$ind)v<- voom(dge, design, plot=TRUE)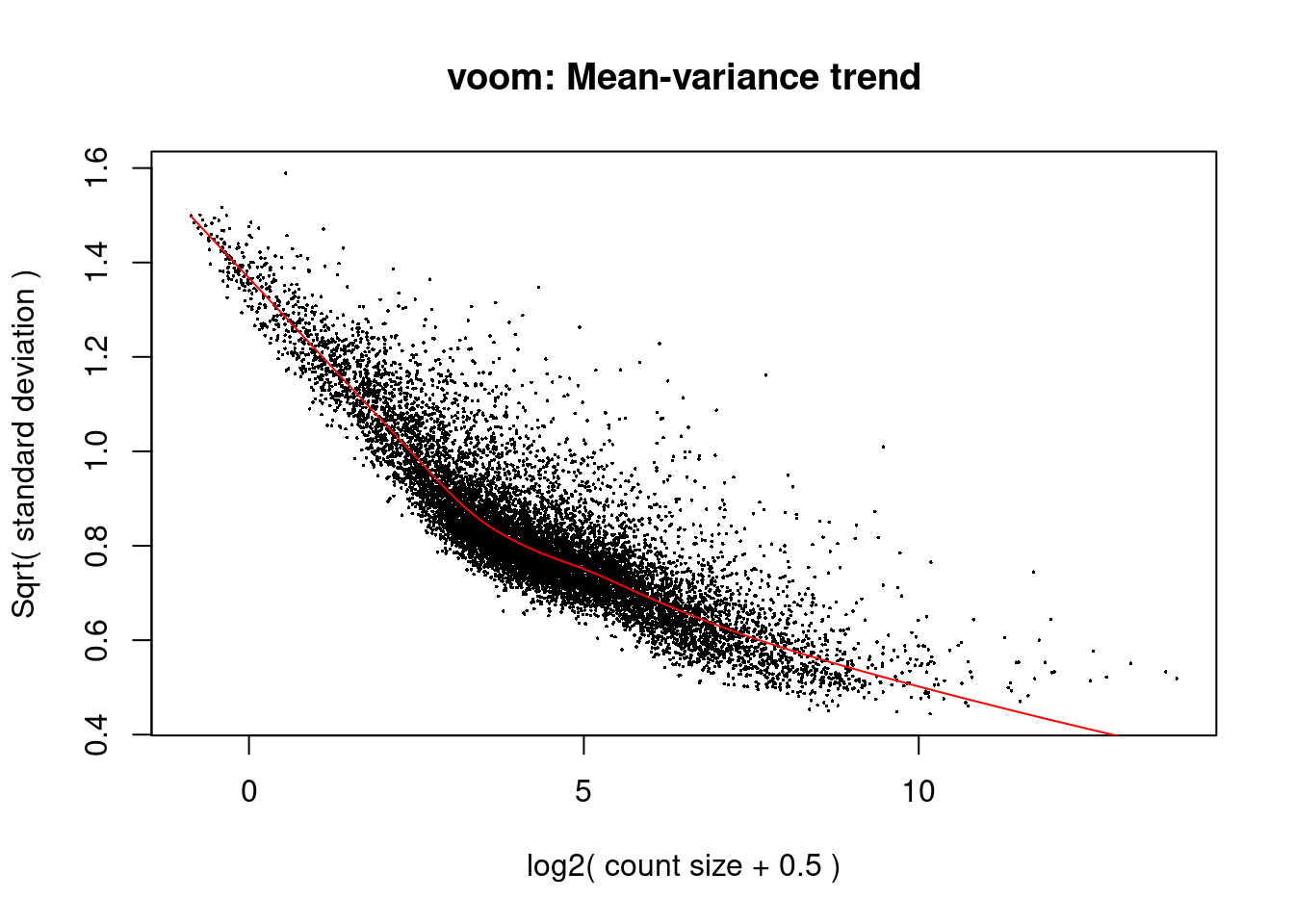
| Version | Author | Date |
|---|---|---|
| 475b623 | KLRhodes | 2021-07-05 |
vAn object of class "EList"
$targets
group lib.size norm.factors cluster batch ind
0.Batch1.SNG-NA18511 1 370114.2 1.048690 0 Batch1 NA18511
0.Batch1.SNG-NA18858 1 56442538.2 1.027369 0 Batch1 NA18858
0.Batch1.SNG-NA19160 1 348812.5 1.023451 0 Batch1 NA19160
0.Batch2.SNG-NA18511 1 596563.8 1.010377 0 Batch2 NA18511
0.Batch2.SNG-NA18858 1 46312654.9 1.024163 0 Batch2 NA18858
Group
0.Batch1.SNG-NA18511 0.Batch1.SNG-NA18511
0.Batch1.SNG-NA18858 0.Batch1.SNG-NA18858
0.Batch1.SNG-NA19160 0.Batch1.SNG-NA19160
0.Batch2.SNG-NA18511 0.Batch2.SNG-NA18511
0.Batch2.SNG-NA18858 0.Batch2.SNG-NA18858
181 more rows ...
$E
0.Batch1.SNG-NA18511 0.Batch1.SNG-NA18858 0.Batch1.SNG-NA19160
SAMD11 3.241309 1.284577 2.104435
NOC2L 6.662772 6.613439 6.541840
PLEKHN1 2.018916 1.857246 2.841400
HES4 6.161874 5.005054 3.326827
ISG15 5.188841 5.670886 5.945737
0.Batch2.SNG-NA18511 0.Batch2.SNG-NA18858 0.Batch2.SNG-NA19160
SAMD11 2.552604 0.9980469 1.722066
NOC2L 6.154640 6.1875509 6.114384
PLEKHN1 2.915174 1.8632702 1.722066
HES4 6.590739 4.4757942 5.266387
ISG15 6.188193 5.4859086 6.114384
0.Batch3.SNG-NA18511 0.Batch3.SNG-NA18858 0.Batch3.SNG-NA19160
SAMD11 1.982800 0.8621931 3.051976
NOC2L 6.309997 6.0462551 6.159935
PLEKHN1 2.051513 1.5239524 2.597410
HES4 5.958403 4.8664805 4.831207
ISG15 6.375117 5.4751883 6.256151
1.Batch1.SNG-NA18511 1.Batch1.SNG-NA18858 1.Batch1.SNG-NA19160
SAMD11 2.841102 3.8411821 2.813415
NOC2L 6.390336 6.0288091 6.266329
PLEKHN1 -1.640025 -0.9137054 -2.520485
HES4 9.406114 8.0140725 8.842737
ISG15 5.338527 5.9566593 5.600184
1.Batch2.SNG-NA18511 1.Batch2.SNG-NA18858 1.Batch2.SNG-NA19160
SAMD11 2.800996 2.954125 2.6056768
NOC2L 6.070414 5.909579 6.1581024
PLEKHN1 -5.159006 -2.255328 -0.7367154
HES4 9.276860 7.676886 8.5476956
ISG15 5.671509 5.816134 5.6383240
1.Batch3.SNG-NA18511 1.Batch3.SNG-NA18858 1.Batch3.SNG-NA19160
SAMD11 2.091638 3.5454125 4.054280
NOC2L 5.977234 5.9732741 6.095721
PLEKHN1 -2.831194 -0.8469049 -3.294448
HES4 8.800741 7.2977533 7.664829
ISG15 5.767479 5.5960386 5.806214
10.Batch1.SNG-NA18511 10.Batch1.SNG-NA18858 10.Batch1.SNG-NA19160
SAMD11 5.000305 1.4589001 1.520778
NOC2L 7.147146 6.5518870 6.164634
PLEKHN1 2.192950 0.3947698 1.520778
HES4 2.192950 5.6183328 7.498058
ISG15 6.280413 5.2253570 5.427669
10.Batch2.SNG-NA18511 10.Batch2.SNG-NA18858 10.Batch2.SNG-NA19160
SAMD11 3.563703 -0.3085178 3.4123508
NOC2L 5.948366 6.0601871 6.2647936
PLEKHN1 2.715706 0.1389412 0.2424258
HES4 6.324515 5.8668617 5.7970146
ISG15 5.347974 5.0297121 4.6347432
10.Batch3.SNG-NA18511 10.Batch3.SNG-NA18858 10.Batch3.SNG-NA19160
SAMD11 3.3598762 -2.218288 4.6721722
NOC2L 6.5465769 6.285537 6.5763122
PLEKHN1 0.6969111 1.241143 -0.4571108
HES4 5.9063645 5.281558 3.6303521
ISG15 5.5548921 6.134858 5.6514137
11.Batch1.SNG-NA18511 11.Batch1.SNG-NA18858 11.Batch1.SNG-NA19160
SAMD11 4.1405344 3.745626 3.1669731
NOC2L 6.5947103 6.468092 6.6742773
PLEKHN1 -0.5033218 1.423698 -0.2630148
HES4 6.6765873 5.947260 6.7531745
ISG15 4.0202402 6.849962 2.7568848
11.Batch2.SNG-NA18511 11.Batch2.SNG-NA18858 11.Batch2.SNG-NA19160
SAMD11 0.02382719 5.952372 2.7671245
NOC2L 5.45009195 5.215406 6.1514314
PLEKHN1 0.02382719 3.630444 0.8002914
HES4 6.79201152 6.437799 6.2804390
ISG15 4.11129003 3.630444 2.5503131
11.Batch3.SNG-NA18511 11.Batch3.SNG-NA18858 11.Batch3.SNG-NA19160
SAMD11 3.489481 2.290705 4.0098056
NOC2L 6.017860 5.750136 6.2254406
PLEKHN1 1.374004 2.290705 -2.7316614
HES4 5.343631 3.875667 5.5398016
ISG15 4.543929 3.875667 0.9687783
12.Batch1.SNG-NA18511 12.Batch1.SNG-NA18858 12.Batch1.SNG-NA19160
SAMD11 3.1948017 1.999870 3.514391
NOC2L 6.2745289 6.523432 5.893605
PLEKHN1 -0.2646299 1.999870 -1.204427
HES4 9.5824274 8.507665 9.281067
ISG15 6.7240548 5.906761 6.321746
12.Batch2.SNG-NA18511 12.Batch2.SNG-NA18858 12.Batch2.SNG-NA19160
SAMD11 3.605991 3.886861 3.454778
NOC2L 6.238260 3.886861 5.671096
PLEKHN1 1.284063 3.886861 -2.217647
HES4 8.701916 8.410423 8.910992
ISG15 5.371526 6.694216 5.776707
12.Batch3.SNG-NA18511 12.Batch3.SNG-NA18858 12.Batch3.SNG-NA19160
SAMD11 2.7005929 3.580428 4.206195
NOC2L 5.4230589 5.902356 5.842007
PLEKHN1 0.3786648 3.580428 -2.009742
HES4 7.8462704 8.335316 8.076394
ISG15 6.0510902 6.750353 6.117823
13.Batch1.SNG-NA18511 13.Batch1.SNG-NA18858 13.Batch1.SNG-NA19160
SAMD11 5.886003 4.5739745 5.864797
NOC2L 6.629905 6.7331731 6.315190
PLEKHN1 -2.073998 0.6670839 -1.168626
HES4 8.235478 5.4219714 8.067389
ISG15 4.771492 5.4219714 6.125995
13.Batch2.SNG-NA18511 13.Batch2.SNG-NA18858 13.Batch2.SNG-NA19160
SAMD11 5.236521 1.388527 4.7197149
NOC2L 6.245779 6.032383 6.4315895
PLEKHN1 -1.042703 1.388527 -0.1382661
HES4 7.982437 6.032383 7.5127856
ISG15 4.964792 5.295417 5.3535870
13.Batch3.SNG-NA18511 13.Batch3.SNG-NA18858 13.Batch3.SNG-NA19160
SAMD11 4.918803 3.295180 6.344438
NOC2L 6.085540 5.410658 5.965285
PLEKHN1 -2.319601 1.710218 -1.469343
HES4 7.440287 5.169650 7.111857
ISG15 4.735681 5.169650 4.834437
14.Batch1.SNG-NA18511 14.Batch1.SNG-NA18858 14.Batch1.SNG-NA19160
SAMD11 5.060818 2.811209 3.995383
NOC2L 6.283211 6.270640 3.995383
PLEKHN1 1.890893 2.811209 3.995383
HES4 4.212821 2.811209 3.995383
ISG15 4.698248 5.133137 3.995383
14.Batch2.SNG-NA18511 14.Batch2.SNG-NA18858 14.Batch2.SNG-NA19160
SAMD11 2.957221 3.677755 5.468826
NOC2L 6.230240 5.999683 6.530754
PLEKHN1 2.957221 3.677755 1.957864
HES4 4.916579 6.485109 5.127789
ISG15 5.515216 5.999683 5.127789
14.Batch3.SNG-NA18511 14.Batch3.SNG-NA18858 14.Batch3.SNG-NA19160
SAMD11 3.211750 2.913119 4.944532
NOC2L 6.481903 5.235047 6.408075
PLEKHN1 1.519074 4.498082 1.898520
HES4 5.745654 6.372551 5.149257
ISG15 5.738468 5.235047 5.574085
15.Batch1.SNG-NA18511 15.Batch1.SNG-NA18858 15.Batch1.SNG-NA19160
SAMD11 3.368292 4.645503 2.732250
NOC2L 6.573407 5.867895 6.819713
PLEKHN1 1.783330 3.060540 2.732250
HES4 3.368292 3.060540 4.317212
ISG15 5.555919 5.382468 5.539605
15.Batch2.SNG-NA18511 15.Batch2.SNG-NA18858 15.Batch2.SNG-NA19160
SAMD11 3.120653 1.574634 4.3878984
NOC2L 6.152131 5.822561 5.9530977
PLEKHN1 2.492622 1.574634 0.3581511
HES4 3.425508 5.034065 3.2967505
ISG15 5.442581 5.481524 5.5413729
15.Batch3.SNG-NA18511 15.Batch3.SNG-NA18858 15.Batch3.SNG-NA19160
SAMD11 1.518668 1.716612 3.636686
NOC2L 6.204768 4.886537 6.121632
PLEKHN1 1.674787 3.301575 2.104673
HES4 4.623005 3.301575 2.777862
ISG15 5.541670 4.038540 5.852321
16.Batch1.SNG-NA18511 16.Batch1.SNG-NA18858 16.Batch1.SNG-NA19160
SAMD11 5.724717 4.3840412 3.256825
NOC2L 6.256828 6.2145809 5.578753
PLEKHN1 -1.394224 0.9540533 3.256825
HES4 8.290524 7.6154356 6.957265
ISG15 5.113570 5.0225331 4.841788
16.Batch2.SNG-NA18511 16.Batch2.SNG-NA18858 16.Batch2.SNG-NA19160
SAMD11 5.039702 3.8693989 3.736211
NOC2L 5.862824 5.9270642 4.958603
PLEKHN1 1.339262 -0.9482244 2.151249
HES4 8.773890 7.0774689 7.009230
ISG15 1.339262 5.8252445 5.321174
16.Batch3.SNG-NA18511 16.Batch3.SNG-NA18858 16.Batch3.SNG-NA19160
SAMD11 2.164585 0.5930146 3.338666
NOC2L 5.334510 6.1476034 4.561058
PLEKHN1 2.164585 0.5930146 1.753703
HES4 7.837010 7.0359581 7.039106
ISG15 6.071475 6.7015391 4.075631
17.Batch1.SNG-NA18511 17.Batch1.SNG-NA18858 17.Batch1.SNG-NA19160
SAMD11 2.297964 4.7769730 1.335469
NOC2L 6.062713 6.0470622 6.171169
PLEKHN1 -2.456924 0.6895102 -1.834456
HES4 9.355655 7.5350002 8.678285
ISG15 5.869506 6.1157749 5.845024
17.Batch2.SNG-NA18511 17.Batch2.SNG-NA18858 17.Batch2.SNG-NA19160
SAMD11 4.198969 6.200073 4.099834
NOC2L 5.783932 6.200073 6.421762
PLEKHN1 4.198969 3.878145 4.099834
HES4 7.368894 3.878145 6.421762
ISG15 4.198969 3.878145 4.099834
18.Batch1.SNG-NA18511 18.Batch1.SNG-NA18858 18.Batch1.SNG-NA19160
SAMD11 2.820019 2.677102 4.289382
NOC2L 5.989944 5.303885 5.667894
PLEKHN1 5.141947 4.964678 5.786214
HES4 8.129875 6.977656 6.858225
ISG15 5.076359 8.187692 6.128446
18.Batch2.SNG-NA18511 18.Batch2.SNG-NA18858 18.Batch2.SNG-NA19160
SAMD11 5.905555 3.906898 4.370176
NOC2L 5.168589 5.805019 5.927393
PLEKHN1 4.431624 4.695394 4.285287
HES4 6.119680 7.188934 6.440565
ISG15 6.471152 8.084091 4.795482
18.Batch3.SNG-NA18511 18.Batch3.SNG-NA18858 18.Batch3.SNG-NA19160
SAMD11 4.917741 2.983232 5.417320
NOC2L 5.424701 5.957237 5.506130
PLEKHN1 4.129245 5.385330 3.963602
HES4 4.917741 6.996056 6.009896
ISG15 4.370254 8.340784 5.272930
19.Batch1.SNG-NA18511 19.Batch1.SNG-NA18858 19.Batch1.SNG-NA19160
SAMD11 2.39453060 3.106031 1.7213752
NOC2L 6.09497032 5.427959 5.7744865
PLEKHN1 0.07260251 3.106031 -0.6005529
HES4 7.29177103 5.913386 6.9919041
ISG15 9.71826094 5.427959 8.2035781
19.Batch2.SNG-NA18511 19.Batch2.SNG-NA19160 19.Batch3.SNG-NA18511
SAMD11 0.2745058 -0.8028146 -0.02751661
NOC2L 6.1571489 5.7970982 5.58719323
PLEKHN1 0.2745058 -0.8028146 -0.02751661
HES4 6.1073958 6.8625213 7.02776583
ISG15 8.1010543 7.5236149 8.07577120
19.Batch3.SNG-NA19160 2.Batch1.SNG-NA18511 2.Batch1.SNG-NA18858
SAMD11 0.80477389 1.945268 2.979981
NOC2L 5.72125053 6.400983 6.219447
PLEKHN1 0.06780829 -1.460725 1.395019
HES4 6.46583937 9.150116 7.372299
ISG15 8.39723092 5.777680 5.591416
2.Batch1.SNG-NA19160 2.Batch2.SNG-NA18511 2.Batch2.SNG-NA18858
SAMD11 3.261891 1.610965 2.8179644
NOC2L 6.177932 6.152919 5.7991872
PLEKHN1 -1.481073 -2.734810 -0.4550541
HES4 8.793628 8.837101 7.0553804
ISG15 6.027602 5.926969 6.0944097
2.Batch2.SNG-NA19160 2.Batch3.SNG-NA18511 2.Batch3.SNG-NA18858
SAMD11 2.206137 2.107558 2.278166
NOC2L 6.167929 6.104215 6.100168
PLEKHN1 -1.990260 -1.424937 -0.529189
HES4 8.470878 8.215567 6.905439
ISG15 5.703227 5.882752 6.341176
2.Batch3.SNG-NA19160 20.Batch1.SNG-NA18858 20.Batch1.SNG-NA19160
SAMD11 4.107743 4.7505948 5.858525
NOC2L 6.383787 6.6588786 6.595490
PLEKHN1 -2.737747 0.9639984 4.273562
HES4 7.681160 5.4364862 4.273562
ISG15 6.429671 5.2714269 7.080917
20.Batch2.SNG-NA18511 20.Batch2.SNG-NA18858 20.Batch2.SNG-NA19160
SAMD11 3.697987 4.288701 3.744458
NOC2L 6.019915 6.054236 6.551813
PLEKHN1 3.697987 1.966773 5.329420
HES4 5.282950 5.267943 3.744458
ISG15 5.282950 5.309165 6.066386
20.Batch3.SNG-NA18511 20.Batch3.SNG-NA18858 20.Batch3.SNG-NA19160
SAMD11 4.111277 3.542864 4.780142
NOC2L 6.379766 5.779904 5.697680
PLEKHN1 2.973774 -1.586419 1.610217
HES4 6.143699 5.071793 3.932145
ISG15 5.509827 5.100082 5.310657
21.Batch1.SNG-NA18858 21.Batch2.SNG-NA18858 21.Batch3.SNG-NA18858
SAMD11 1.9422668 0.357488 2.589616
NOC2L 6.2880416 6.288225 6.049048
PLEKHN1 0.3573043 0.357488 0.267688
HES4 8.3056715 8.305855 8.239232
ISG15 5.4865873 5.486771 5.759541
3.Batch1.SNG-NA18511 3.Batch1.SNG-NA18858 3.Batch1.SNG-NA19160
SAMD11 3.021604 1.157502 2.229045
NOC2L 6.928494 6.613193 6.752607
PLEKHN1 1.436641 1.361516 2.229045
HES4 3.758569 4.351573 3.814007
ISG15 3.021604 4.981715 5.688477
3.Batch2.SNG-NA18511 3.Batch2.SNG-NA18858 3.Batch2.SNG-NA19160
SAMD11 2.773094 -0.4967796 1.022343
NOC2L 6.559690 6.1395950 5.545905
PLEKHN1 2.036128 1.7641804 2.607305
HES4 4.538629 4.5536320 3.344271
ISG15 4.151605 5.2198329 5.109806
3.Batch3.SNG-NA18511 3.Batch3.SNG-NA18858 3.Batch3.SNG-NA19160
SAMD11 2.592464 0.4858941 4.0950205
NOC2L 6.537323 6.2560299 6.4928974
PLEKHN1 1.958592 0.7839754 0.4705296
HES4 5.852155 4.8301172 4.8628470
ISG15 6.022452 5.3919961 5.5437786
4.Batch1.SNG-NA18511 4.Batch1.SNG-NA18858 4.Batch1.SNG-NA19160
SAMD11 2.9204865 3.386559 3.229835
NOC2L 6.2406753 6.556484 6.256673
PLEKHN1 0.4523376 1.801596 -1.174920
HES4 5.5141138 5.997993 5.938349
ISG15 6.4568390 6.755793 6.629505
4.Batch2.SNG-NA18511 4.Batch2.SNG-NA18858 4.Batch2.SNG-NA19160
SAMD11 3.2788062 4.432943 2.9963374
NOC2L 5.7403181 5.570447 5.9798493
PLEKHN1 -0.3765456 2.111015 -0.2220861
HES4 4.9810064 3.695978 5.2093312
ISG15 6.4839206 6.198478 6.3289566
4.Batch3.SNG-NA18511 4.Batch3.SNG-NA18858 4.Batch3.SNG-NA19160
SAMD11 3.00126940 2.351456 3.320616
NOC2L 6.04713641 5.810888 6.147016
PLEKHN1 -0.01503241 2.351456 -1.257652
HES4 3.83841892 5.158811 4.932172
ISG15 6.25515671 6.875018 6.415559
5.Batch1.SNG-NA18511 5.Batch1.SNG-NA18858 5.Batch1.SNG-NA19160
SAMD11 4.6773674 4.1943542 3.082059
NOC2L 6.4205919 6.6175656 6.257231
PLEKHN1 0.1538054 0.7349226 -1.263715
HES4 7.0963199 6.8434470 6.623675
ISG15 6.0686888 4.8223854 4.908434
5.Batch2.SNG-NA18511 5.Batch2.SNG-NA18858 5.Batch2.SNG-NA19160
SAMD11 5.2614065 2.701928 3.310124
NOC2L 6.2466095 5.871853 6.177288
PLEKHN1 -0.3533033 2.701928 -1.444764
HES4 5.9865467 5.509283 6.540509
ISG15 5.9504774 5.023856 4.784055
5.Batch3.SNG-NA18511 5.Batch3.SNG-NA18858 5.Batch3.SNG-NA19160
SAMD11 2.4440258 1.965729 4.469679
NOC2L 6.1181142 6.720617 6.036594
PLEKHN1 -0.0584745 1.965729 -1.953226
HES4 4.5063101 5.872620 5.596751
ISG15 4.3789308 5.135654 4.014864
6.Batch1.SNG-NA18511 6.Batch1.SNG-NA18858 6.Batch1.SNG-NA19160
SAMD11 3.701322 1.812091 2.0190301
NOC2L 6.134282 6.288201 5.6435210
PLEKHN1 2.964357 1.340022 0.4340676
HES4 6.588848 5.517683 4.1345074
ISG15 5.286285 5.202810 6.0487775
6.Batch2.SNG-NA18511 6.Batch2.SNG-NA18858 6.Batch2.SNG-NA19160
SAMD11 3.196284 1.051967 3.863615
NOC2L 6.300621 5.913374 6.109091
PLEKHN1 2.459319 1.664306 1.231346
HES4 5.122284 5.618152 4.975507
ISG15 4.333788 5.387922 5.729597
6.Batch3.SNG-NA18511 6.Batch3.SNG-NA18858 6.Batch3.SNG-NA19160
SAMD11 2.674473 0.8356805 2.8916891
NOC2L 6.332004 5.8146351 6.0768005
PLEKHN1 2.369619 1.2594883 0.8212998
HES4 5.595039 5.3157565 4.7661582
ISG15 5.782002 5.5562793 5.7939924
7.Batch1.SNG-NA18511 7.Batch1.SNG-NA18858 7.Batch1.SNG-NA19160
SAMD11 4.04667493 4.462000 2.2510454
NOC2L 6.21962128 6.068179 6.0715517
PLEKHN1 -0.09628303 1.292075 -0.7124287
HES4 8.51227983 5.345187 6.5067398
ISG15 5.59487888 5.738332 5.8695250
7.Batch2.SNG-NA18511 7.Batch2.SNG-NA18858 7.Batch2.SNG-NA19160
SAMD11 2.966143 3.191139 2.7256477
NOC2L 5.983057 5.639467 5.8116715
PLEKHN1 -1.176815 1.459956 0.5434444
HES4 6.936928 5.560934 4.9876629
ISG15 5.198225 6.153722 5.5118897
7.Batch3.SNG-NA18511 7.Batch3.SNG-NA18858 7.Batch3.SNG-NA19160
SAMD11 2.709869 2.728992 2.750957
NOC2L 5.943616 5.839416 6.114173
PLEKHN1 -4.110310 2.076916 -1.466274
HES4 5.367448 5.152204 5.927760
ISG15 5.701867 6.212075 5.806123
8.Batch1.SNG-NA18511 8.Batch1.SNG-NA18858 8.Batch1.SNG-NA19160
SAMD11 4.7405193 2.792468 4.536257
NOC2L 6.0353113 5.828092 5.985868
PLEKHN1 0.8690341 5.514934 3.424364
HES4 7.1013460 6.879931 7.073794
ISG15 6.9926852 6.845579 7.565565
8.Batch2.SNG-NA18511 8.Batch2.SNG-NA18858 8.Batch2.SNG-NA19160
SAMD11 4.602152 4.427559 3.813348
NOC2L 5.965072 5.345097 5.796070
PLEKHN1 1.155265 3.579562 1.491420
HES4 7.167025 6.683899 6.780341
ISG15 6.805975 6.012522 7.155740
8.Batch3.SNG-NA18511 8.Batch3.SNG-NA18858 8.Batch3.SNG-NA19160
SAMD11 4.9890423 3.364356 4.4278206
NOC2L 6.0370895 5.842403 5.9934502
PLEKHN1 0.7179385 2.141963 0.1507957
HES4 5.7144663 7.243501 6.4090958
ISG15 6.2772420 7.271246 7.0978434
9.Batch1.SNG-NA18511 9.Batch1.SNG-NA18858 9.Batch1.SNG-NA19160
SAMD11 2.714479 2.6340280 3.0230135
NOC2L 5.920249 5.7093162 5.9864876
PLEKHN1 -3.013441 -0.1733269 -0.8838771
HES4 9.152408 7.7982167 8.6623743
ISG15 4.911371 4.9559561 4.9823715
9.Batch2.SNG-NA18511 9.Batch2.SNG-NA18858 9.Batch2.SNG-NA19160
SAMD11 2.918695 2.404528 1.559203
NOC2L 5.982064 5.445555 5.633550
PLEKHN1 -3.767805 -1.295912 -2.141237
HES4 9.033701 7.117716 8.395011
ISG15 5.240623 4.853835 4.824548
9.Batch3.SNG-NA18511 9.Batch3.SNG-NA18858 9.Batch3.SNG-NA19160
SAMD11 2.115656 3.5459682 3.919656
NOC2L 5.722038 6.0961653 5.794125
PLEKHN1 -3.169746 0.7386133 -1.572197
HES4 8.481754 6.5715033 7.605222
ISG15 5.740147 5.9480667 5.370317
11151 more rows ...
$weights
[,1] [,2] [,3] [,4] [,5] [,6] [,7]
[1,] 0.4450400 7.635886 0.4441243 0.5420162 6.296748 0.4623990 2.658602
[2,] 3.3137030 31.340461 3.1169616 3.7923968 26.032900 3.1948249 10.782539
[3,] 0.3431286 11.064648 0.3236890 0.5203411 10.662923 0.4185935 2.265241
[4,] 2.6232635 20.978728 2.1903115 2.9777454 17.933651 2.3200401 7.764601
[5,] 2.6791839 28.354751 2.6297823 3.0354365 24.077544 2.7446417 9.281377
[,8] [,9] [,10] [,11] [,12] [,13] [,14]
[1,] 6.532047 1.680222 6.8808189 1.2462676 5.5734398 6.9884634 2.1852199
[2,] 27.832522 7.162742 16.7110603 4.6086262 13.0940906 16.9688713 6.6046475
[3,] 9.745134 1.151128 0.5191766 0.1987132 0.3595256 0.6814088 0.2872754
[4,] 17.331689 4.418269 38.0363080 9.4115927 26.2793112 39.5709524 13.1070625
[5,] 27.283193 6.345082 13.5454160 4.1876624 11.0612652 14.0223983 6.1692539
[,15] [,16] [,17] [,18] [,19] [,20] [,21]
[1,] 4.110474 5.328326 0.9558655 3.6789182 0.2285108 2.454820 0.3174851
[2,] 10.146396 13.779335 4.1097994 9.5394835 1.9059374 10.241660 2.3881962
[3,] 0.299085 0.367176 0.1987132 0.2195747 0.1987132 2.235076 0.1987132
[4,] 20.535830 28.409948 7.8882636 17.4778497 1.2439211 7.124591 1.4389522
[5,] 8.711483 12.045859 4.0845100 8.6585607 0.9200965 8.083426 1.4176066
[,22] [,23] [,24] [,25] [,26] [,27] [,28]
[1,] 0.4527023 2.768475 0.5093337 0.8776575 1.377799 0.7181283 1.6180628
[2,] 3.1416619 11.551637 3.1747427 4.9779759 7.301735 4.0985398 4.2218474
[3,] 0.2222991 2.919790 0.2328009 0.3166808 1.203314 0.2569441 0.2683915
[4,] 2.6181885 8.276047 2.3842199 3.5072526 4.475191 2.7160705 4.5668097
[5,] 2.2168870 9.340755 2.3611182 3.5296708 6.129707 3.0983614 1.6288450
[,29] [,30] [,31] [,32] [,33] [,34] [,35]
[1,] 0.3875742 4.769047 0.9655449 0.1987132 3.2668430 1.0761175 0.2241332
[2,] 2.1171690 10.614608 3.1757610 0.5135507 7.6547264 3.4733437 1.1256336
[3,] 0.1987132 1.226156 0.2295005 0.1987132 0.8428438 0.2084229 0.1987132
[4,] 1.9122153 10.280932 3.4861011 0.4735843 7.5651167 3.4332622 0.8404230
[5,] 0.5963882 4.686600 1.0143976 0.1987132 3.2878058 1.3591290 0.3842181
[,36] [,37] [,38] [,39] [,40] [,41] [,42]
[1,] 3.2819522 1.4538719 0.3051236 3.8731757 0.4971728 0.1987132 2.9300098
[2,] 7.9401383 3.4570803 1.2971879 7.9163343 1.8667190 0.3854202 5.9489936
[3,] 0.6739192 0.2353306 0.1987132 0.7961459 0.1987132 0.1987132 0.6332065
[4,] 7.0769245 10.1036385 3.8371518 18.7926568 5.7345556 1.8905378 14.7758313
[5,] 3.6418507 3.6479300 1.8403356 8.6586554 2.0645795 0.5351278 6.7075603
[,43] [,44] [,45] [,46] [,47] [,48] [,49]
[1,] 0.7855584 0.1987132 4.544835 4.7090371 1.2869941 3.5078187 5.1467010
[2,] 2.6654920 0.4576278 9.377223 7.5909544 2.8421608 5.3957216 8.2208096
[3,] 0.1987132 0.1987132 1.072123 0.3215796 0.1987132 0.2134359 0.4523639
[4,] 7.0769986 1.9144733 20.580159 10.6564952 3.4534651 7.0340483 11.7355728
[5,] 2.9786995 0.7141464 11.026154 4.8655250 2.1368956 3.5458047 5.4441528
[,50] [,51] [,52] [,53] [,54] [,55] [,56]
[1,] 0.6970509 2.3522889 4.5225136 0.5677310 3.438252 0.5627378 0.2709818
[2,] 2.0137235 3.3712948 7.5712782 1.7556918 5.483315 1.7607278 0.8873523
[3,] 0.1987132 0.1987132 0.3127185 0.1987132 0.212890 0.2394857 0.2651483
[4,] 2.6233673 4.5666784 9.8530067 2.1291071 6.565708 1.1939299 0.4898359
[5,] 1.2691571 2.4348274 5.2981364 1.1970068 3.945911 0.9479857 0.6416178
[,57] [,58] [,59] [,60] [,61] [,62] [,63]
[1,] 0.2190964 1.6307454 0.1987132 1.8517660 5.954114 0.2226100 4.240491
[2,] 0.4865768 3.2882210 0.4707231 3.3256993 11.305171 0.7231184 7.942820
[3,] 0.1987132 0.7197171 0.1987132 0.7659782 3.222663 0.2228563 2.336645
[4,] 0.3065156 2.8006380 0.2908456 2.5766496 8.646628 0.3632512 5.296146
[5,] 0.3194294 2.5105404 0.3675046 2.6447480 9.006497 0.6064738 6.490996
[,64] [,65] [,66] [,67] [,68] [,69] [,70]
[1,] 0.8605106 0.1987132 0.2524759 1.4701187 0.2719767 1.7904303 3.870092
[2,] 3.2825024 0.8167145 1.0453101 4.4528701 1.6892313 4.6871800 10.688469
[3,] 0.3772345 0.1987132 0.1987132 0.7501816 0.3670085 0.8508985 2.431147
[4,] 1.5382270 0.2397901 0.3028021 2.3804772 0.4253954 2.2267923 5.127481
[5,] 2.4878864 0.6084577 0.6464284 3.2480595 1.2716855 3.5649320 8.613556
[,71] [,72] [,73] [,74] [,75] [,76] [,77]
[1,] 0.2494383 3.337072 2.6349144 2.801507 0.2608948 0.5407937 3.497105
[2,] 1.5981352 8.863604 5.0047677 6.197829 0.6157899 1.7287404 7.934208
[3,] 0.2778857 1.963777 0.4995893 1.582310 0.1987132 0.1987132 2.639266
[4,] 0.3476321 3.654472 10.1542014 10.690351 1.8452780 3.7936333 13.653012
[5,] 1.3450097 7.403318 3.4769682 5.068681 0.3922165 0.9588502 6.711661
[,78] [,79] [,80] [,81] [,82] [,83] [,84]
[1,] 0.3757713 0.3496720 0.565541 0.4432272 3.852832 0.7196186 3.2043068
[2,] 0.9756948 1.0674222 2.258859 1.2862576 8.224433 2.7294286 6.4840130
[3,] 0.1987132 0.1987132 0.290463 0.1987132 1.011975 0.3508244 0.6882352
[4,] 2.5927445 2.7084243 3.554398 2.6835567 12.216237 3.5193556 8.8819219
[5,] 0.6115266 0.6802651 1.967046 0.8694749 5.285207 1.9872249 4.2719508
[,85] [,86] [,87] [,88] [,89] [,90] [,91]
[1,] 0.1987132 0.1987132 0.1987132 2.255475 2.626460 2.994465 0.7716062
[2,] 0.4306776 0.4571883 0.4271990 3.439242 4.672462 4.472757 1.7912729
[3,] 0.1987132 0.1987132 0.1987132 2.163528 4.445678 2.814695 0.9499859
[4,] 0.9098870 0.7520584 0.7455137 5.466875 6.289282 6.285757 2.9339354
[5,] 0.2409321 0.3075050 0.2535475 3.917241 6.174146 5.340878 2.1816874
[,92] [,93] [,94] [,95] [,96] [,97] [,98]
[1,] 0.8719899 2.411550 1.059479 0.9908655 2.196467 0.3184286 0.1987132
[2,] 2.3337395 3.433928 2.352606 2.5895982 3.265070 3.1107878 0.6708436
[3,] 2.5920305 2.598523 1.024431 2.4348302 2.043340 0.1987132 0.1987132
[4,] 3.0692228 4.989004 3.203817 3.0624077 4.208898 4.5874628 0.9936914
[5,] 3.0203557 4.205849 2.839020 3.5289492 4.252005 6.0302057 2.4115488
[,99] [,100] [,101] [,102] [,103] [,104] [,105]
[1,] 0.4565656 0.2547825 0.4345693 0.2834095 0.6093320 4.0934800 0.6177052
[2,] 3.7368979 2.7127593 3.6113842 2.9803319 4.7385532 12.5757704 3.6460613
[3,] 0.2570808 0.1987132 0.3016413 0.1987132 0.3375632 0.5055052 0.1987132
[4,] 4.9760583 3.9320849 4.9304425 3.9637640 5.7340453 24.5327839 6.7411546
[5,] 7.5853721 5.1944609 7.5142231 6.2332408 10.0383221 10.8782784 3.5793319
[,106] [,107] [,108] [,109] [,110] [,111] [,112]
[1,] 5.7191748 4.4534292 1.5568759 3.5121772 4.2280373 0.6110106 2.702567
[2,] 15.7220453 13.4977017 6.2911527 10.4596937 13.3259952 3.7589824 8.222391
[3,] 0.7911966 0.7350772 0.4089625 0.4834342 0.5442522 0.1987132 0.262186
[4,] 28.0637719 27.0429223 11.1245152 18.6945764 24.0876829 6.3695184 13.383520
[5,] 14.1543887 11.9074471 6.3205611 9.5971373 12.4194473 4.0395443 7.966697
[,113] [,114] [,115] [,116] [,117] [,118] [,119]
[1,] 2.779204 0.2406968 0.2614043 2.117486 0.2653932 1.2567421 2.679236
[2,] 5.686117 0.4960839 0.6094028 4.269177 0.5591422 2.8495673 5.637212
[3,] 1.965183 0.1987132 0.1987132 1.630669 0.1987132 0.2970333 1.893786
[4,] 3.451358 0.2697882 0.3873056 2.777347 0.3083522 1.8405168 3.157892
[5,] 4.780359 0.3364840 0.3918664 3.654995 0.3869957 2.3099983 5.175885
[,120] [,121] [,122] [,123] [,124] [,125] [,126]
[1,] 0.7407660 0.8190154 0.3121558 0.3167106 0.2662622 4.161359 0.1987132
[2,] 1.9724846 5.5315466 2.9267274 3.0355623 2.5735036 19.234231 1.7924980
[3,] 0.1987132 0.3782213 0.2023535 0.1987132 0.1987132 5.508282 0.1987132
[4,] 0.7915303 10.8995333 6.1754206 5.7898051 1.1269430 10.220617 0.5553208
[5,] 1.4607881 4.2674035 2.4035467 2.6573915 1.1015247 13.149311 0.6991330
[,127] [,128] [,129] [,130] [,131] [,132] [,133]
[1,] 0.3629642 4.023765 0.2900037 2.188462 4.070766 0.8325587 4.547791
[2,] 3.1176489 18.715577 2.5739362 9.681284 19.498893 5.2317397 10.984090
[3,] 0.2882532 6.247102 0.2216492 1.354568 5.492370 0.4756096 1.079983
[4,] 1.7869899 10.159512 0.9618049 5.187201 9.597349 2.5180142 8.930320
[5,] 1.7497761 13.057100 1.2519957 6.093866 14.353595 3.2864565 11.494405
[,134] [,135] [,136] [,137] [,138] [,139] [,140]
[1,] 0.6949837 7.012769 3.1535107 0.2400486 4.964338 4.0271499 0.2119745
[2,] 3.0692971 15.056053 7.9799158 1.2094303 11.162821 10.2457564 1.0689692
[3,] 0.2897443 2.074209 0.7583684 0.1987132 1.563016 0.9069737 0.1987132
[4,] 2.1578692 11.141324 6.5312576 0.6445506 8.389085 7.6785723 0.4902416
[5,] 3.7184240 16.413863 8.5389420 1.7971839 12.400404 11.5539992 1.7870325
[,141] [,142] [,143] [,144] [,145] [,146] [,147]
[1,] 5.796735 2.6628332 0.6352631 7.37093 1.4586756 0.2174281 5.321564
[2,] 13.134665 6.2837241 2.8427010 15.21156 3.8791878 0.9694245 11.428039
[3,] 1.599079 0.3079723 0.2001782 1.55936 0.2077496 0.1987132 1.133905
[4,] 8.990701 6.5212870 2.5826774 14.26048 4.1395735 0.8266982 10.938476
[5,] 15.418144 3.6295175 1.9181584 9.96560 2.4498733 0.5362878 7.563226
[,148] [,149] [,150] [,151] [,152] [,153] [,154]
[1,] 2.5480764 0.2908001 6.208027 0.3540901 3.022380 0.5915734 0.3899860
[2,] 6.1960548 1.5790312 13.460556 2.3302600 10.946010 2.9835488 2.5010295
[3,] 0.2951409 0.1987132 1.163395 0.1987132 3.209517 0.2644561 0.2387404
[4,] 5.9007757 1.0878445 11.706562 1.9353259 8.329838 2.3700291 2.1798087
[5,] 3.9139638 0.9172054 9.494676 1.5106921 9.511755 2.3800065 1.7683249
[,155] [,156] [,157] [,158] [,159] [,160] [,161]
[1,] 5.000355 1.2060722 1.6338733 3.590832 1.5062916 2.4868042 1.3692439
[2,] 16.983451 4.5031672 5.9261657 13.285488 5.2852982 6.1609768 4.5843548
[3,] 6.298963 0.5953182 0.6634174 3.944910 0.5773386 0.3015331 0.3812588
[4,] 13.248909 3.4900822 4.6294370 9.413761 3.6532819 7.0144846 4.4854899
[5,] 15.051336 3.4995383 4.7183312 12.453277 4.4000197 5.1745782 4.4693680
[,162] [,163] [,164] [,165] [,166] [,167] [,168]
[1,] 3.8565532 3.1010917 2.949831 3.6168040 4.8401607 0.9085295 4.4435448
[2,] 9.1508364 7.9555696 8.519472 8.6730482 12.1082264 3.7582965 10.6342025
[3,] 0.5716856 0.5507444 1.388239 0.6658699 0.8988023 0.2883334 0.7290225
[4,] 9.3198751 9.1568439 8.542115 9.0251838 12.5958715 3.3670839 10.0355165
[5,] 8.1621483 6.9059123 8.505602 7.8900434 11.2202908 4.0109907 10.2578990
[,169] [,170] [,171] [,172] [,173] [,174] [,175]
[1,] 4.730062 1.0600924 3.3433816 5.339768 0.5697367 4.883732 4.785093
[2,] 8.032094 2.7363507 5.4432788 8.942418 1.8239950 7.777743 8.382837
[3,] 1.804972 0.5879815 0.9232076 2.505375 0.4267903 2.191645 1.886155
[4,] 11.658797 3.4337041 7.3791088 13.195649 2.5578369 10.473683 11.264470
[5,] 10.020032 3.9240611 7.2719828 11.335706 2.8449640 10.320828 11.252554
[,176] [,177] [,178] [,179] [,180] [,181] [,182]
[1,] 0.8119817 8.968680 3.3154081 0.6709907 2.9010075 3.8810454 1.0841725
[2,] 2.4699975 13.963861 8.9828393 3.2173461 7.3143403 10.3077094 4.2874347
[3,] 0.4793855 3.404313 0.2855301 0.1987132 0.2221273 0.4383226 0.2401836
[4,] 2.8816805 16.962175 19.5527602 6.7883007 14.5260881 22.9998603 8.9992459
[5,] 3.7994910 19.502292 6.7529034 2.8253024 5.6566129 7.9864277 3.7112519
[,183] [,184] [,185] [,186]
[1,] 2.4103863 3.1165301 0.3521328 1.8678162
[2,] 5.9371342 8.7179503 2.2712719 4.9509839
[3,] 0.2073898 0.2672415 0.1987132 0.1987132
[4,] 12.3204123 17.5690780 4.1486468 9.5698669
[5,] 4.6508224 7.1047874 2.0859660 4.1204997
11151 more rows ...
$design
dge$samples$cluster0 dge$samples$cluster1 dge$samples$cluster10
1 1 0 0
2 1 0 0
3 1 0 0
4 1 0 0
5 1 0 0
dge$samples$cluster11 dge$samples$cluster12 dge$samples$cluster13
1 0 0 0
2 0 0 0
3 0 0 0
4 0 0 0
5 0 0 0
dge$samples$cluster14 dge$samples$cluster15 dge$samples$cluster16
1 0 0 0
2 0 0 0
3 0 0 0
4 0 0 0
5 0 0 0
dge$samples$cluster17 dge$samples$cluster18 dge$samples$cluster19
1 0 0 0
2 0 0 0
3 0 0 0
4 0 0 0
5 0 0 0
dge$samples$cluster2 dge$samples$cluster20 dge$samples$cluster21
1 0 0 0
2 0 0 0
3 0 0 0
4 0 0 0
5 0 0 0
dge$samples$cluster3 dge$samples$cluster4 dge$samples$cluster5
1 0 0 0
2 0 0 0
3 0 0 0
4 0 0 0
5 0 0 0
dge$samples$cluster6 dge$samples$cluster7 dge$samples$cluster8
1 0 0 0
2 0 0 0
3 0 0 0
4 0 0 0
5 0 0 0
dge$samples$cluster9 dge$samples$batchBatch2 dge$samples$batchBatch3
1 0 0 0
2 0 0 0
3 0 0 0
4 0 1 0
5 0 1 0
dge$samples$indNA18858 dge$samples$indNA19160
1 0 0
2 1 0
3 0 1
4 0 0
5 1 0
181 more rows ...fit<- lmFit(v,design)nclust<- length(unique(dge$samples$cluster))
contrasts<- NULL
for (i in 1:nclust){
c<- c(rep(-1,nclust),0,0,0,0)
c[i]<- nclust-1
contrasts<- cbind(contrasts, c)
}contrasts c c c c c c c c c c c c c c c c c c c c c c
[1,] 21 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1
[2,] -1 21 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1
[3,] -1 -1 21 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1
[4,] -1 -1 -1 21 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1
[5,] -1 -1 -1 -1 21 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1
[6,] -1 -1 -1 -1 -1 21 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1
[7,] -1 -1 -1 -1 -1 -1 21 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1
[8,] -1 -1 -1 -1 -1 -1 -1 21 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1
[9,] -1 -1 -1 -1 -1 -1 -1 -1 21 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1
[10,] -1 -1 -1 -1 -1 -1 -1 -1 -1 21 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1
[11,] -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 21 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1
[12,] -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 21 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1
[13,] -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 21 -1 -1 -1 -1 -1 -1 -1 -1 -1
[14,] -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 21 -1 -1 -1 -1 -1 -1 -1 -1
[15,] -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 21 -1 -1 -1 -1 -1 -1 -1
[16,] -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 21 -1 -1 -1 -1 -1 -1
[17,] -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 21 -1 -1 -1 -1 -1
[18,] -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 21 -1 -1 -1 -1
[19,] -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 21 -1 -1 -1
[20,] -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 21 -1 -1
[21,] -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 21 -1
[22,] -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 21
[23,] 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
[24,] 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
[25,] 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
[26,] 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0#first, testing all pairwise cluster comparisons.
fit<- contrasts.fit(fit, contrasts= contrasts)efit<- eBayes(fit)plotSA(efit)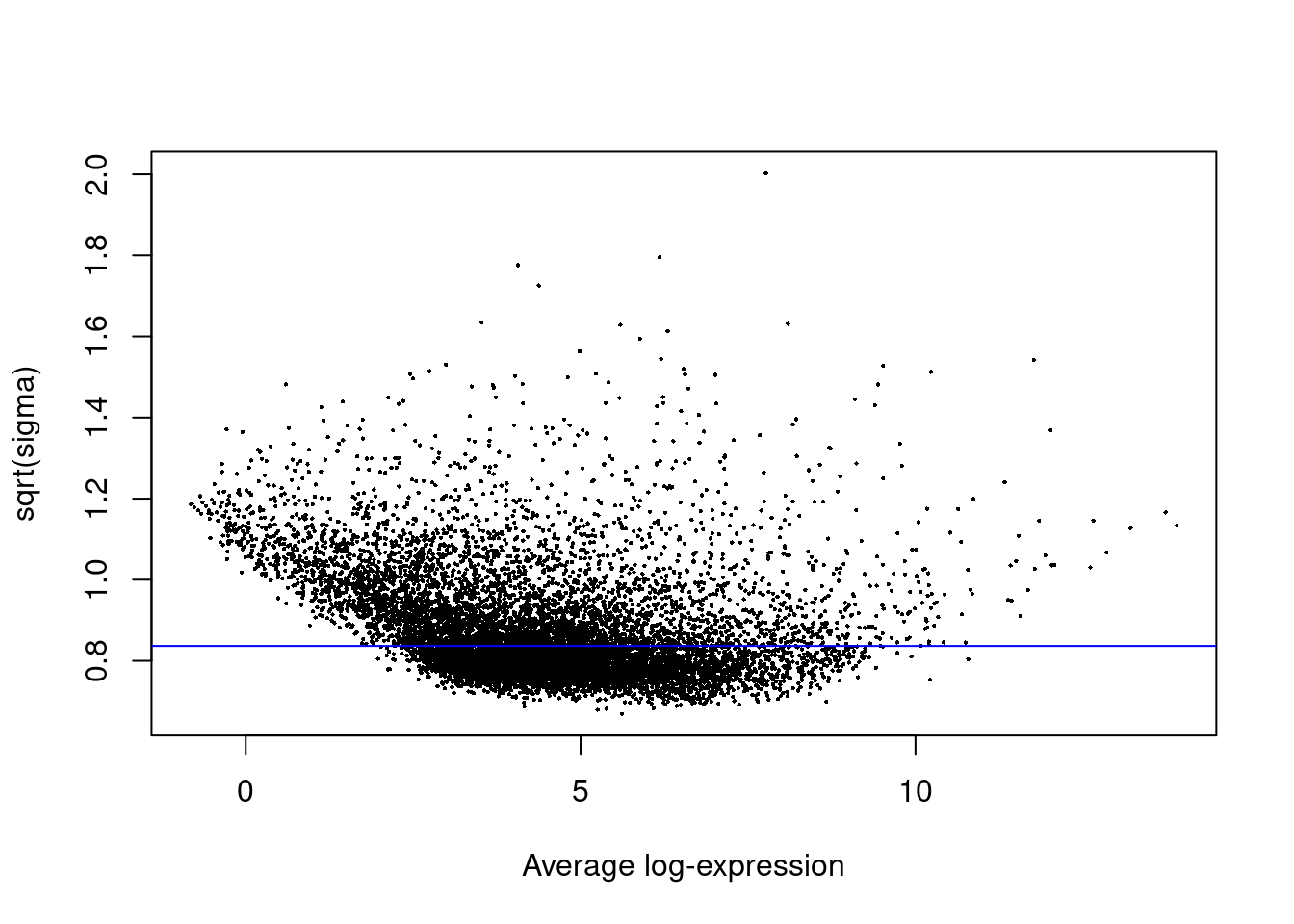
| Version | Author | Date |
|---|---|---|
| 475b623 | KLRhodes | 2021-07-05 |
summary(decideTests(efit)) c c c c c c c c c c c c c c
Down 3632 3869 2932 3016 2354 3868 3101 2939 1751 1824 2690 2317 3866 1901
NotSig 3285 3820 5163 5294 6486 3820 4361 4694 7478 7209 5274 6066 3866 6430
Up 4239 3467 3061 2846 2316 3468 3694 3523 1927 2123 3192 2773 3424 2825
c c c c c c c c
Down 644 3459 2919 2854 3145 1456 2912 3201
NotSig 8933 3877 4958 5603 4314 7469 4835 4637
Up 1579 3820 3279 2699 3697 2231 3409 3318output.list<- list()
for (i in 1:nclust){
ta<- topTable(efit, coef=i,n=nrow(fit))
output.list[[i]]<- ta
}
listnames<- c(paste0("Cluster", unique(dge$samples$cluster)))
names(output.list)<- as.vector(listnames)#add cluster number to colnames
for (i in 1:nclust){
d<- names(output.list[i])
colnames(output.list[[i]])<- paste(d, colnames(output.list[[i]]), sep=".")
}
all<- left_join(rownames_to_column(output.list[[1]]), rownames_to_column(output.list[[2]]), by= c("rowname" = "rowname"))
for (i in 3:nclust){
all<- left_join(all, rownames_to_column(output.list[[i]]), by= c("rowname" = "rowname"))
}
colnames(all)[1]<- "gene"
write.csv(all, "/project2/gilad/katie/Pilot_HumanEBs/Embryoid_Body_Pilot_Workflowr/output/NEW_Pseudobulk_Limma_res0.8_OnevAllTopTables.csv")sessionInfo()R version 3.6.1 (2019-07-05)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Scientific Linux 7.4 (Nitrogen)
Matrix products: default
BLAS/LAPACK: /software/openblas-0.2.19-el7-x86_64/lib/libopenblas_haswellp-r0.2.19.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] tibble_3.1.5 edgeR_3.28.1 limma_3.42.2 dplyr_1.0.2
[5] Matrix_1.2-18 Seurat_3.2.0 workflowr_1.6.2
loaded via a namespace (and not attached):
[1] Rtsne_0.15 colorspace_2.0-2 deldir_0.1-28
[4] ellipsis_0.3.2 ggridges_0.5.2 rprojroot_2.0.2
[7] fs_1.4.2 spatstat.data_1.4-3 leiden_0.3.3
[10] listenv_0.8.0 npsurv_0.4-0 ggrepel_0.9.0
[13] fansi_0.5.0 codetools_0.2-16 splines_3.6.1
[16] lsei_1.2-0 knitr_1.29 polyclip_1.10-0
[19] jsonlite_1.7.2 ica_1.0-2 cluster_2.1.0
[22] png_0.1-7 uwot_0.1.10 shiny_1.5.0
[25] sctransform_0.2.1 compiler_3.6.1 httr_1.4.2
[28] fastmap_1.0.1 lazyeval_0.2.2 later_1.1.0.1
[31] htmltools_0.5.0 tools_3.6.1 rsvd_1.0.3
[34] igraph_1.2.6 gtable_0.3.0 glue_1.4.2
[37] RANN_2.6.1 reshape2_1.4.4 rappdirs_0.3.3
[40] Rcpp_1.0.6 spatstat_1.64-1 vctrs_0.3.8
[43] ape_5.4-1 nlme_3.1-140 lmtest_0.9-37
[46] xfun_0.16 stringr_1.4.0 globals_0.12.5
[49] mime_0.9 miniUI_0.1.1.1 lifecycle_1.0.1
[52] irlba_2.3.3 goftest_1.2-2 future_1.18.0
[55] MASS_7.3-51.4 zoo_1.8-8 scales_1.1.1
[58] promises_1.1.1 spatstat.utils_1.17-0 parallel_3.6.1
[61] RColorBrewer_1.1-2 yaml_2.2.1 reticulate_1.20
[64] pbapply_1.4-2 gridExtra_2.3 ggplot2_3.3.5
[67] rpart_4.1-15 stringi_1.5.3 highr_0.8
[70] rlang_0.4.11 pkgconfig_2.0.3 evaluate_0.14
[73] lattice_0.20-38 ROCR_1.0-11 purrr_0.3.4
[76] tensor_1.5 patchwork_1.1.1 htmlwidgets_1.5.1
[79] cowplot_1.1.1 tidyselect_1.1.0 here_0.1-11
[82] RcppAnnoy_0.0.18 plyr_1.8.6 magrittr_2.0.1
[85] R6_2.5.1 generics_0.1.0 pillar_1.6.3
[88] whisker_0.4 mgcv_1.8-28 fitdistrplus_1.0-14
[91] survival_3.2-3 abind_1.4-5 future.apply_1.6.0
[94] crayon_1.4.1 KernSmooth_2.23-15 utf8_1.2.2
[97] plotly_4.9.2.1 rmarkdown_2.3 locfit_1.5-9.4
[100] grid_3.6.1 data.table_1.13.4 git2r_0.26.1
[103] digest_0.6.28 xtable_1.8-4 tidyr_1.1.0
[106] httpuv_1.5.4 munsell_0.5.0 viridisLite_0.4.0
sessionInfo()R version 3.6.1 (2019-07-05)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Scientific Linux 7.4 (Nitrogen)
Matrix products: default
BLAS/LAPACK: /software/openblas-0.2.19-el7-x86_64/lib/libopenblas_haswellp-r0.2.19.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] tibble_3.1.5 edgeR_3.28.1 limma_3.42.2 dplyr_1.0.2
[5] Matrix_1.2-18 Seurat_3.2.0 workflowr_1.6.2
loaded via a namespace (and not attached):
[1] Rtsne_0.15 colorspace_2.0-2 deldir_0.1-28
[4] ellipsis_0.3.2 ggridges_0.5.2 rprojroot_2.0.2
[7] fs_1.4.2 spatstat.data_1.4-3 leiden_0.3.3
[10] listenv_0.8.0 npsurv_0.4-0 ggrepel_0.9.0
[13] fansi_0.5.0 codetools_0.2-16 splines_3.6.1
[16] lsei_1.2-0 knitr_1.29 polyclip_1.10-0
[19] jsonlite_1.7.2 ica_1.0-2 cluster_2.1.0
[22] png_0.1-7 uwot_0.1.10 shiny_1.5.0
[25] sctransform_0.2.1 compiler_3.6.1 httr_1.4.2
[28] fastmap_1.0.1 lazyeval_0.2.2 later_1.1.0.1
[31] htmltools_0.5.0 tools_3.6.1 rsvd_1.0.3
[34] igraph_1.2.6 gtable_0.3.0 glue_1.4.2
[37] RANN_2.6.1 reshape2_1.4.4 rappdirs_0.3.3
[40] Rcpp_1.0.6 spatstat_1.64-1 vctrs_0.3.8
[43] ape_5.4-1 nlme_3.1-140 lmtest_0.9-37
[46] xfun_0.16 stringr_1.4.0 globals_0.12.5
[49] mime_0.9 miniUI_0.1.1.1 lifecycle_1.0.1
[52] irlba_2.3.3 goftest_1.2-2 future_1.18.0
[55] MASS_7.3-51.4 zoo_1.8-8 scales_1.1.1
[58] promises_1.1.1 spatstat.utils_1.17-0 parallel_3.6.1
[61] RColorBrewer_1.1-2 yaml_2.2.1 reticulate_1.20
[64] pbapply_1.4-2 gridExtra_2.3 ggplot2_3.3.5
[67] rpart_4.1-15 stringi_1.5.3 highr_0.8
[70] rlang_0.4.11 pkgconfig_2.0.3 evaluate_0.14
[73] lattice_0.20-38 ROCR_1.0-11 purrr_0.3.4
[76] tensor_1.5 patchwork_1.1.1 htmlwidgets_1.5.1
[79] cowplot_1.1.1 tidyselect_1.1.0 here_0.1-11
[82] RcppAnnoy_0.0.18 plyr_1.8.6 magrittr_2.0.1
[85] R6_2.5.1 generics_0.1.0 pillar_1.6.3
[88] whisker_0.4 mgcv_1.8-28 fitdistrplus_1.0-14
[91] survival_3.2-3 abind_1.4-5 future.apply_1.6.0
[94] crayon_1.4.1 KernSmooth_2.23-15 utf8_1.2.2
[97] plotly_4.9.2.1 rmarkdown_2.3 locfit_1.5-9.4
[100] grid_3.6.1 data.table_1.13.4 git2r_0.26.1
[103] digest_0.6.28 xtable_1.8-4 tidyr_1.1.0
[106] httpuv_1.5.4 munsell_0.5.0 viridisLite_0.4.0