ReferenceAnn_DE
Last updated: 2021-07-05
Checks: 5 2
Knit directory: Embryoid_Body_Pilot_Workflowr/analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown is untracked by Git. To know which version of the R Markdown file created these results, you'll want to first commit it to the Git repo. If you're still working on the analysis, you can ignore this warning. When you're finished, you can run wflow_publish to commit the R Markdown file and build the HTML.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it's best to always run the code in an empty environment.
The command set.seed(20200804) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Using absolute paths to the files within your workflowr project makes it difficult for you and others to run your code on a different machine. Change the absolute path(s) below to the suggested relative path(s) to make your code more reproducible.
| absolute | relative |
|---|---|
| /project2/gilad/katie/Pilot_HumanEBs/Embryoid_Body_Pilot_Workflowr/output/mergedObjects/Harmony.Batchindividual.rds | ../output/mergedObjects/Harmony.Batchindividual.rds |
| /project2/gilad/katie/Pilot_HumanEBs/Embryoid_Body_Pilot_Workflowr/output/MostCommonAnnotation.FiveNearestRefCells.csv | ../output/MostCommonAnnotation.FiveNearestRefCells.csv |
| /project2/gilad/katie/Pilot_HumanEBs/Embryoid_Body_Pilot_Workflowr/output/figs/Supp_RefAnnDE_cardiomyocyte.png | ../output/figs/Supp_RefAnnDE_cardiomyocyte.png |
| /project2/gilad/katie/Pilot_HumanEBs/Embryoid_Body_Pilot_Workflowr/output/figs/Supp_RefAnnDE_hepatoblast.png | ../output/figs/Supp_RefAnnDE_hepatoblast.png |
| /project2/gilad/katie/Pilot_HumanEBs/Embryoid_Body_Pilot_Workflowr/output/figs/Supp_RefAnnDE_mesothelial.png | ../output/figs/Supp_RefAnnDE_mesothelial.png |
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version c8767ac. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: analysis/.Rhistory
Ignored: output/.Rhistory
Untracked files:
Untracked: GSE122380_raw_counts.txt.gz
Untracked: UTF1_plots.Rmd
Untracked: analysis/IntegrateReference_SCTregressCaoPlusScHCL_JustEarlyEcto.Rmd
Untracked: analysis/IntegrateReference_SCTregressCaoPlusScHCL_JustEndo.Rmd
Untracked: analysis/IntegrateReference_SCTregressCaoPlusScHCL_JustMeso.Rmd
Untracked: analysis/IntegrateReference_SCTregressCaoPlusScHCL_JustNeuralCrest.Rmd
Untracked: analysis/IntegrateReference_SCTregressCaoPlusScHCL_JustNeuron.Rmd
Untracked: analysis/IntegrateReference_SCTregressCaoPlusScHCL_JustPluri.Rmd
Untracked: analysis/OLD/
Untracked: analysis/Pseudobulk_Limma_Harmony.BatchIndividual_ClusterRes0.8_minPCT0.2.Rmd
Untracked: analysis/Pseudobulk_Limma_Harmony.BatchIndividual_ClusterRes1_minPCT0.2.Rmd
Untracked: analysis/Pseudobulk_VariancePartition_Harmony.Batchindividual_ClusterRes0.1_byCluster.Rmd
Untracked: analysis/RefInt_ComparingFulltoPartialIntegrationAnnotations.Rmd
Untracked: analysis/ReferenceAnn_DE.Rmd
Untracked: analysis/SingleCell_HierarchicalClustering_NoGeneFilter.Rmd
Untracked: analysis/SingleCell_VariancePartitionByCluster_Harmony.Batchindividual_ClusterRes0.1_minPCT0.2.Rmd
Untracked: analysis/VarPartPlots_res0.1_SCT.Rmd
Untracked: analysis/VarPart_SC_res0.1_SCT.Rmd
Untracked: analysis/child/
Untracked: analysis/k10topics_Explore.Rmd
Untracked: analysis/k6topics_Explore.Rmd
Untracked: build_refint_scale.R
Untracked: build_refint_sct.R
Untracked: build_stuff.R
Untracked: build_varpart_sc.R
Untracked: code/.ipynb_checkpoints/
Untracked: code/CellRangerPreprocess.Rmd
Untracked: code/ConvertToDGE.Rmd
Untracked: code/ConvertToDGE_PseudoBulk.Rmd
Untracked: code/ConvertToDGE_SingleCellRes_minPCT0.2.Rmd
Untracked: code/EB.getHumanMetadata.Rmd
Untracked: code/GEO_processed_data.Rmd
Untracked: code/PowerAnalysis_NoiseRatio.ipynb
Untracked: code/Untitled.ipynb
Untracked: code/Untitled1.ipynb
Untracked: code/compile_fits.Rmd
Untracked: code/fit_all_models.sh
Untracked: code/fit_poisson_nmf.R
Untracked: code/fit_poisson_nmf.sbatch
Untracked: code/functions_for_fit_comparison.Rmd
Untracked: code/get_genelist_byPCTthresh.Rmd
Untracked: code/prefit_poisson_nmf.R
Untracked: code/prefit_poisson_nmf.sbatch
Untracked: code/prepare_data_for_fastTopics.Rmd
Untracked: data/HCL_Fig1_adata.h5ad
Untracked: data/HCL_Fig1_adata.h5seurat
Untracked: data/dge/
Untracked: data/dge_raw_data.tar.gz
Untracked: data/ref.expr.rda
Untracked: figure/
Untracked: output/CR_sampleQCrds/
Untracked: output/CaoEtAl.Obj.CellsOfAllClusters.ProteinCodingGenes.rds
Untracked: output/CaoEtAl.Obj.rds
Untracked: output/ClusterInfo_res0.1.csv
Untracked: output/DGELists/
Untracked: output/DownSampleVarPart.rds
Untracked: output/Frequency.MostCommonAnnotation.FiveNearestRefCells.csv
Untracked: output/GEOsubmissionProcessedFiles/
Untracked: output/GeneLists_by_minPCT/
Untracked: output/MostCommonAnnotation.FiveNearestRefCells.csv
Untracked: output/NearestReferenceCell.Cao.hESC.EuclideanDistanceinHarmonySpace.csv
Untracked: output/NearestReferenceCell.Cao.hESC.FrequencyofEachAnnotation.csv
Untracked: output/NearestReferenceCell.SCTregressRNAassay.Cao.hESC.EuclideanDistanceinHarmonySpace.csv
Untracked: output/NearestReferenceCell.SCTregressRNAassay.Cao.hESC.FrequencyofEachAnnotation.csv
Untracked: output/Pseudobulk_Limma_res0.1_OnevAllTopTables.csv
Untracked: output/Pseudobulk_Limma_res0.1_OnevAll_top10Upregby_adjP.csv
Untracked: output/Pseudobulk_Limma_res0.1_OnevAll_top10Upregby_logFC.csv
Untracked: output/Pseudobulk_Limma_res0.5_OnevAllTopTables.csv
Untracked: output/Pseudobulk_Limma_res0.8_OnevAllTopTables.csv
Untracked: output/Pseudobulk_Limma_res1_OnevAllTopTables.csv
Untracked: output/Pseudobulk_VarPart.ByCluster.Res0.1.rds
Untracked: output/ResidualVariances_fromDownSampAnalysis.csv
Untracked: output/SingleCell_VariancePartition_RNA_Res0.1_minPCT0.2.rds
Untracked: output/SingleCell_VariancePartition_Res0.1_minPCT0.2.rds
Untracked: output/SingleCell_VariancePartition_SCT_Res0.1_minPCT0.2.rds
Untracked: output/TopicModelling_k10_top10drivergenes.byBeta.csv
Untracked: output/TopicModelling_k6_top10drivergenes.byBeta.csv
Untracked: output/TopicModelling_k6_top15drivergenes.byZ.csv
Untracked: output/TranferredAnnotations_ReferenceInt_JustEarlyEcto.csv
Untracked: output/TranferredAnnotations_ReferenceInt_JustEndoderm.csv
Untracked: output/TranferredAnnotations_ReferenceInt_JustMeso.csv
Untracked: output/TranferredAnnotations_ReferenceInt_JustNeuralCrest.csv
Untracked: output/TranferredAnnotations_ReferenceInt_JustNeuron.csv
Untracked: output/TranferredAnnotations_ReferenceInt_JustPluripotent.csv
Untracked: output/VarPart.ByCluster.Res0.1.rds
Untracked: output/azimuth/
Untracked: output/downsamp_10800cells_10subreps_medianexplainedbyresiduals_varpart_PsB.rds
Untracked: output/downsamp_16200cells_10subreps_medianexplainedbyresiduals_varpart_PsB.rds
Untracked: output/downsamp_21600cells_10subreps_medianexplainedbyresiduals_varpart_PsB.rds
Untracked: output/downsamp_2700cells_10subreps_medianexplainedbyresiduals_varpart_PsB.rds
Untracked: output/downsamp_2700cells_10subreps_medianexplainedbyresiduals_varpart_scres.rds
Untracked: output/downsamp_5400cells_10subreps_medianexplainedbyresiduals_varpart_PsB.rds
Untracked: output/downsamp_7200cells_10subreps_medianexplainedbyresiduals_varpart_PsB.rds
Untracked: output/fasttopics/
Untracked: output/figs/
Untracked: output/merge.Cao.SCTwRegressOrigIdent.rds
Untracked: output/merge.all.SCTwRegressOrigIdent.Harmony.rds
Untracked: output/merged.SCT.counts.matrix.rds
Untracked: output/merged.raw.counts.matrix.rds
Untracked: output/mergedObjects/
Untracked: output/pdfs/
Untracked: output/sampleQCrds/
Untracked: output/splitgpm_gsea_results/
Untracked: slurm-12005914.out
Untracked: slurm-12005923.out
Unstaged changes:
Deleted: analysis/IntegrateAnalysis.afterFilter.HarmonyBatch.Rmd
Deleted: analysis/IntegrateAnalysis.afterFilter.HarmonyBatchSampleIDindividual.Rmd
Modified: analysis/IntegrateAnalysis.afterFilter.HarmonyBatchindividual.Rmd
Deleted: analysis/IntegrateAnalysis.afterFilter.NOHARMONYjustmerge.Rmd
Deleted: analysis/IntegrateAnalysis.afterFilter.SCTregressBatchIndividual.Rmd
Deleted: analysis/IntegrateAnalysis.afterFilter.SCTregressBatchIndividualHarmonyBatchindividual.Rmd
Modified: analysis/Pseudobulk_HierarchicalClustering_Harmony.Batchindividual_ClusterRes0.1_minPCT0.2.Rmd
Modified: analysis/Pseudobulk_HierarchicalClustering_Harmony.Batchindividual_ClusterRes0.5_minPCT0.2.Rmd
Modified: analysis/Pseudobulk_HierarchicalClustering_Harmony.Batchindividual_ClusterRes0.8_minPCT0.2.Rmd
Modified: analysis/Pseudobulk_HierarchicalClustering_Harmony.Batchindividual_ClusterRes1_minPCT0.2.Rmd
Modified: analysis/Pseudobulk_Limma_Harmony.BatchIndividual_ClusterRes0.1_minPCT0.2.Rmd
Modified: analysis/Pseudobulk_Limma_Harmony.BatchIndividual_ClusterRes0.5_minPCT0.2.Rmd
Modified: analysis/Pseudobulk_VariancePartition_Harmony.Batchindividual_ClusterRes0.1_minPCT0.2.Rmd
Modified: analysis/Pseudobulk_VariancePartition_Harmony.Batchindividual_ClusterRes0.5_minPCT0.2.Rmd
Modified: analysis/Pseudobulk_VariancePartition_Harmony.Batchindividual_ClusterRes0.8_minPCT0.2.Rmd
Modified: analysis/Pseudobulk_VariancePartition_Harmony.Batchindividual_ClusterRes1_minPCT0.2.Rmd
Deleted: analysis/RunscHCL_HarmonyBatchInd.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
There are no past versions. Publish this analysis with wflow_publish() to start tracking its development.
library(Seurat)
library(limma)
library(edgeR)
library(Matrix)
library(dplyr)
Attaching package: 'dplyr'The following objects are masked from 'package:stats':
filter, lagThe following objects are masked from 'package:base':
intersect, setdiff, setequal, unionlibrary(scater)Loading required package: SingleCellExperimentLoading required package: SummarizedExperimentLoading required package: GenomicRangesLoading required package: stats4Loading required package: BiocGenericsLoading required package: parallel
Attaching package: 'BiocGenerics'The following objects are masked from 'package:parallel':
clusterApply, clusterApplyLB, clusterCall, clusterEvalQ,
clusterExport, clusterMap, parApply, parCapply, parLapply,
parLapplyLB, parRapply, parSapply, parSapplyLBThe following objects are masked from 'package:dplyr':
combine, intersect, setdiff, unionThe following object is masked from 'package:Matrix':
whichThe following object is masked from 'package:limma':
plotMAThe following objects are masked from 'package:stats':
IQR, mad, sd, var, xtabsThe following objects are masked from 'package:base':
Filter, Find, Map, Position, Reduce, anyDuplicated, append,
as.data.frame, basename, cbind, colnames, dirname, do.call,
duplicated, eval, evalq, get, grep, grepl, intersect, is.unsorted,
lapply, mapply, match, mget, order, paste, pmax, pmax.int, pmin,
pmin.int, rank, rbind, rownames, sapply, setdiff, sort, table,
tapply, union, unique, unsplit, which, which.max, which.minLoading required package: S4Vectors
Attaching package: 'S4Vectors'The following objects are masked from 'package:dplyr':
first, renameThe following object is masked from 'package:Matrix':
expandThe following object is masked from 'package:base':
expand.gridLoading required package: IRanges
Attaching package: 'IRanges'The following objects are masked from 'package:dplyr':
collapse, desc, sliceLoading required package: GenomeInfoDbLoading required package: BiobaseWelcome to Bioconductor
Vignettes contain introductory material; view with
'browseVignettes()'. To cite Bioconductor, see
'citation("Biobase")', and for packages 'citation("pkgname")'.Loading required package: DelayedArrayLoading required package: matrixStats
Attaching package: 'matrixStats'The following objects are masked from 'package:Biobase':
anyMissing, rowMediansThe following object is masked from 'package:dplyr':
countLoading required package: BiocParallel
Attaching package: 'DelayedArray'The following objects are masked from 'package:matrixStats':
colMaxs, colMins, colRanges, rowMaxs, rowMins, rowRangesThe following objects are masked from 'package:base':
aperm, apply, rowsum
Attaching package: 'SummarizedExperiment'The following object is masked from 'package:Seurat':
Assays
Attaching package: 'SingleCellExperiment'The following object is masked from 'package:edgeR':
cpmLoading required package: ggplot2
Attaching package: 'scater'The following object is masked from 'package:limma':
plotMDSmerged<- readRDS("/project2/gilad/katie/Pilot_HumanEBs/Embryoid_Body_Pilot_Workflowr/output/mergedObjects/Harmony.Batchindividual.rds")#load annotations
ann<- read.csv("/project2/gilad/katie/Pilot_HumanEBs/Embryoid_Body_Pilot_Workflowr/output/MostCommonAnnotation.FiveNearestRefCells.csv")merged@meta.data$RefAnn<- ann$AnnotationDimPlot(merged, group.by= "RefAnn")+NoLegend()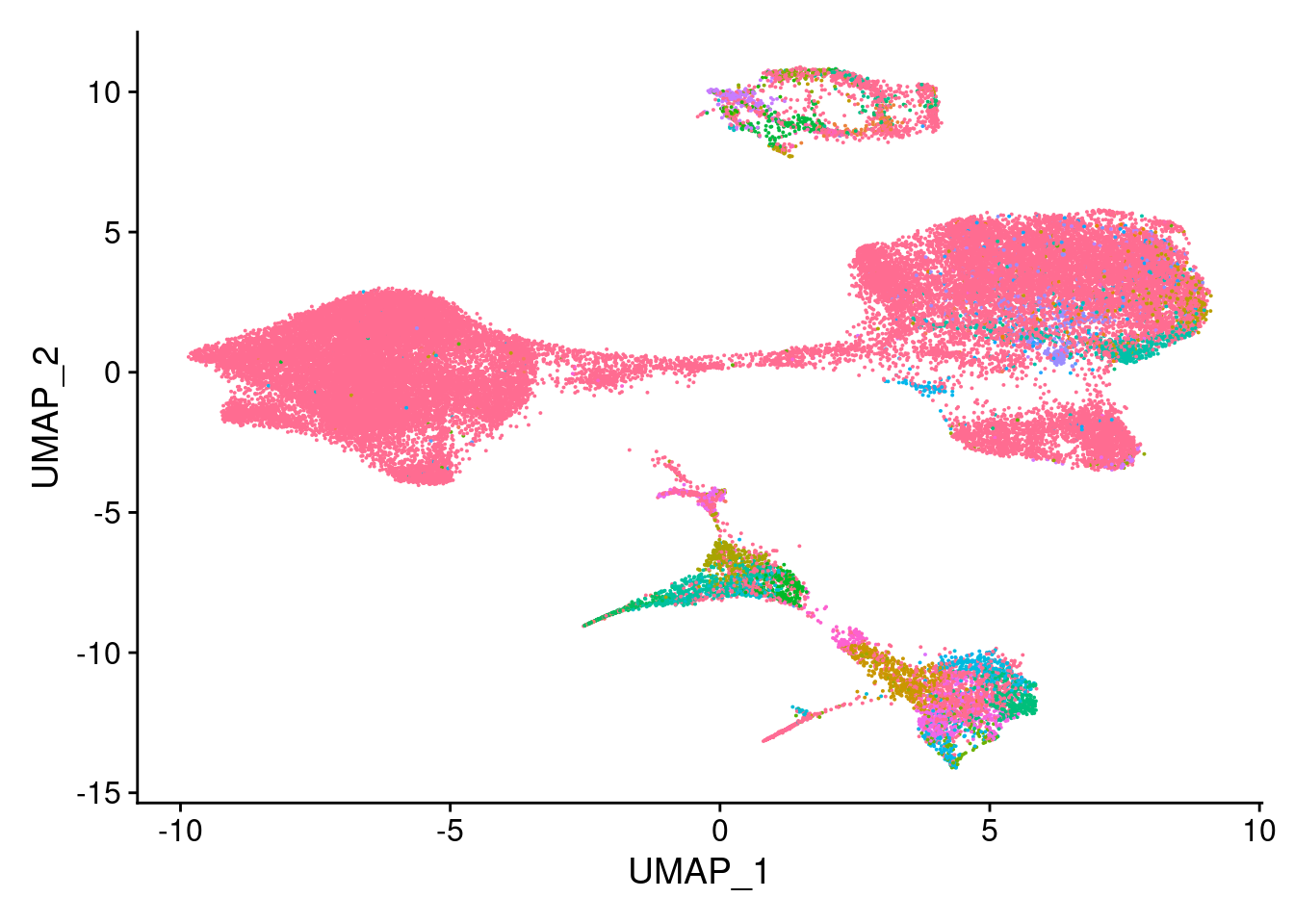
clusts<- (unique(merged@meta.data$RefAnn))
#theres 1 thymocyte and it doesnt want to plot it. so removing
clusts<- clusts[-57]
#or myeloid
clusts<- clusts[-58]
#or excitatory neurons
clusts<- clusts[-66]
#or antigen presenting
clusts<- clusts[-65]
plts<- NULL
for (i in 1:length(clusts)){
b<- clusts[i]
plts[[i]]<- DimPlot(merged, cells= c(rownames(merged@meta.data[merged@meta.data$RefAnn == as.character(b),])))+NoLegend()+ggtitle(paste0(b))
}
plts[[1]]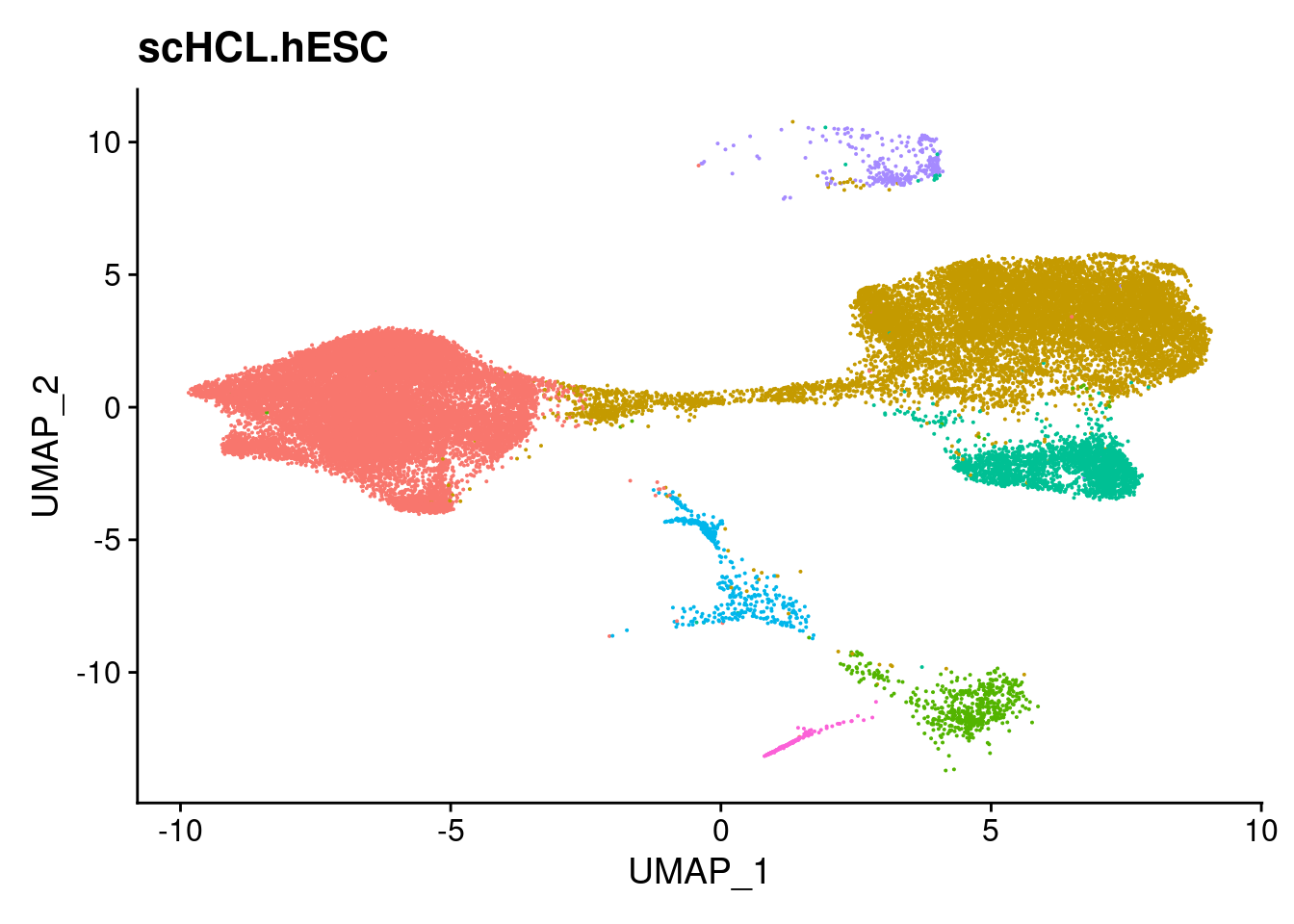
[[2]]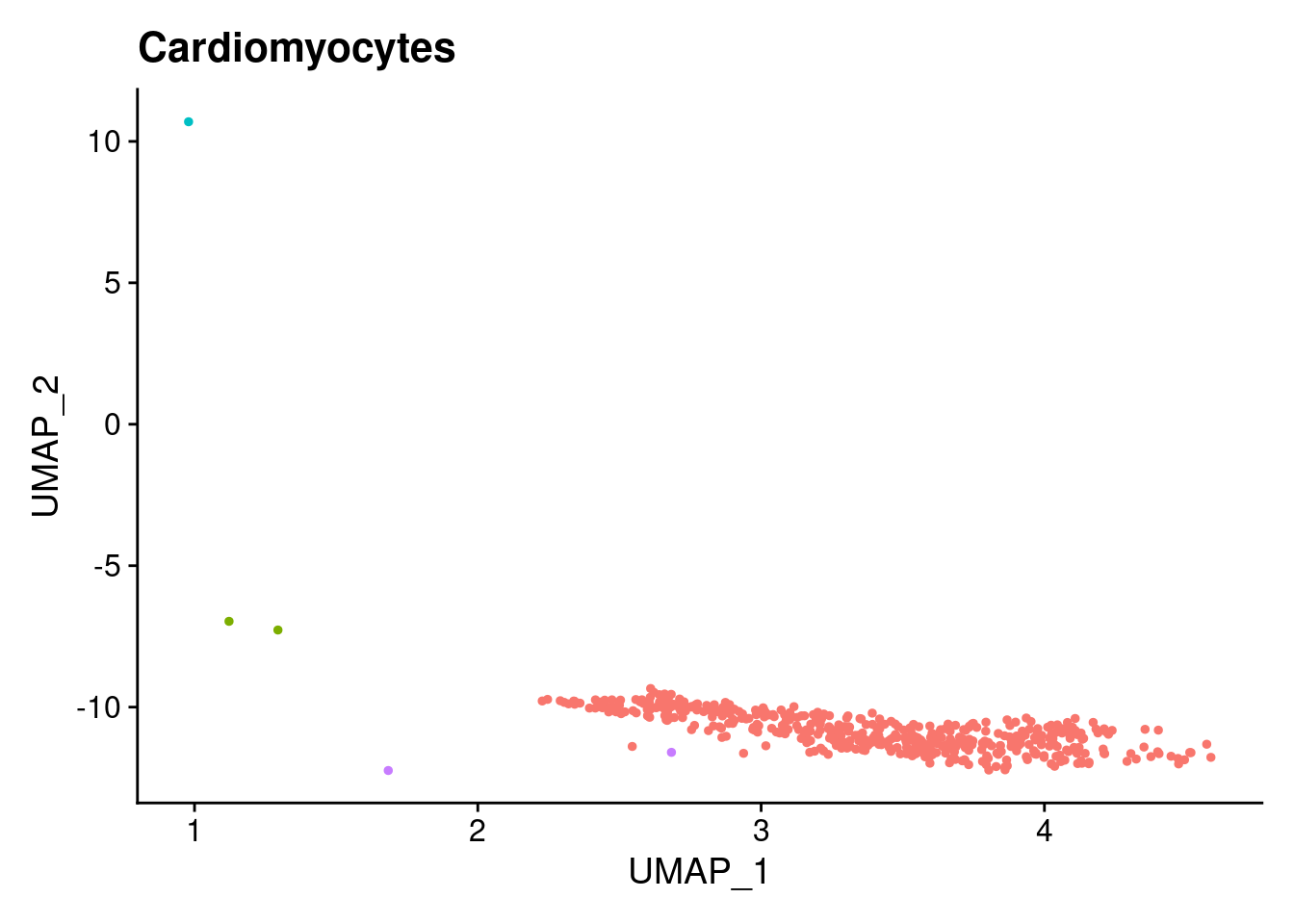
[[3]]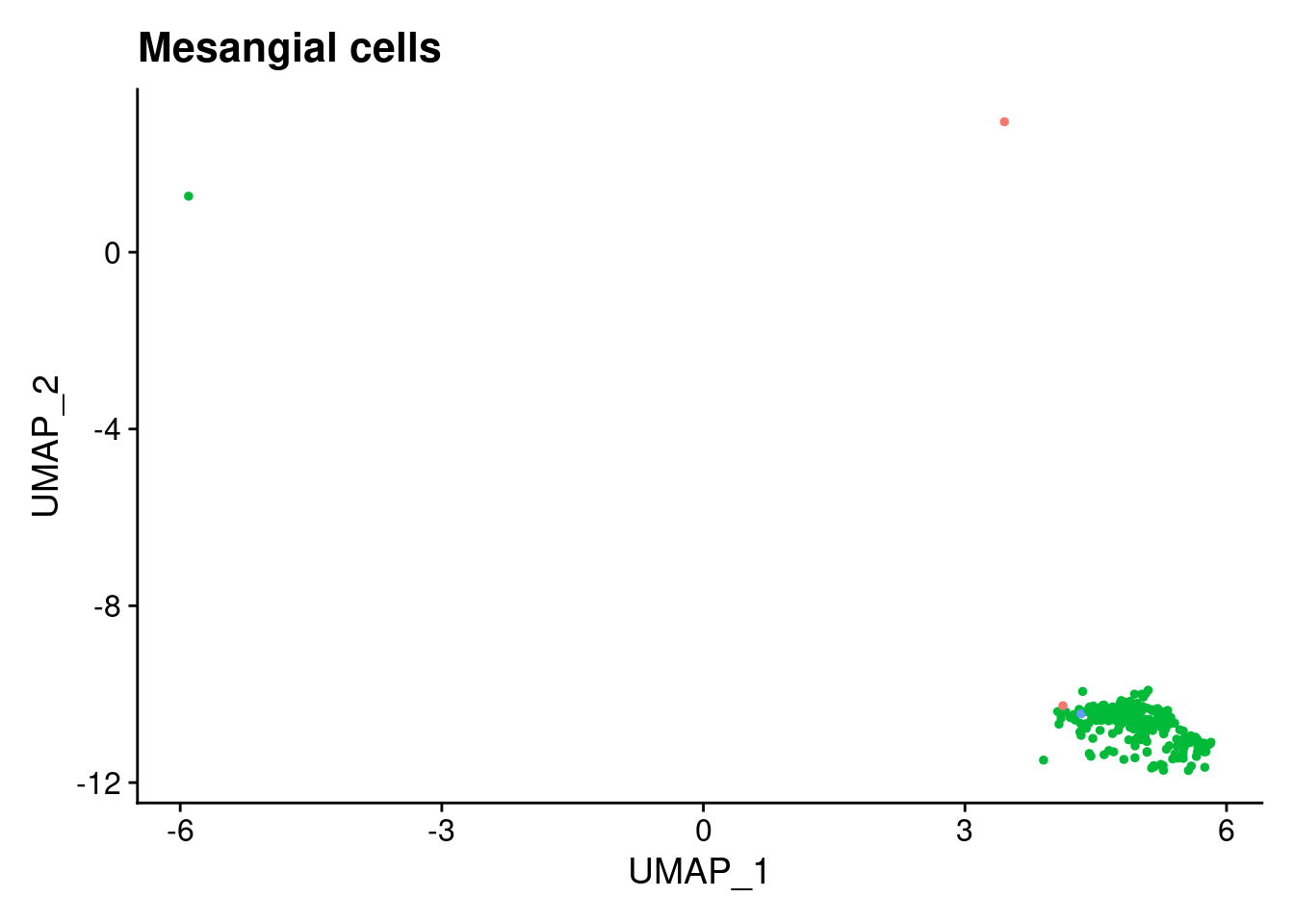
[[4]]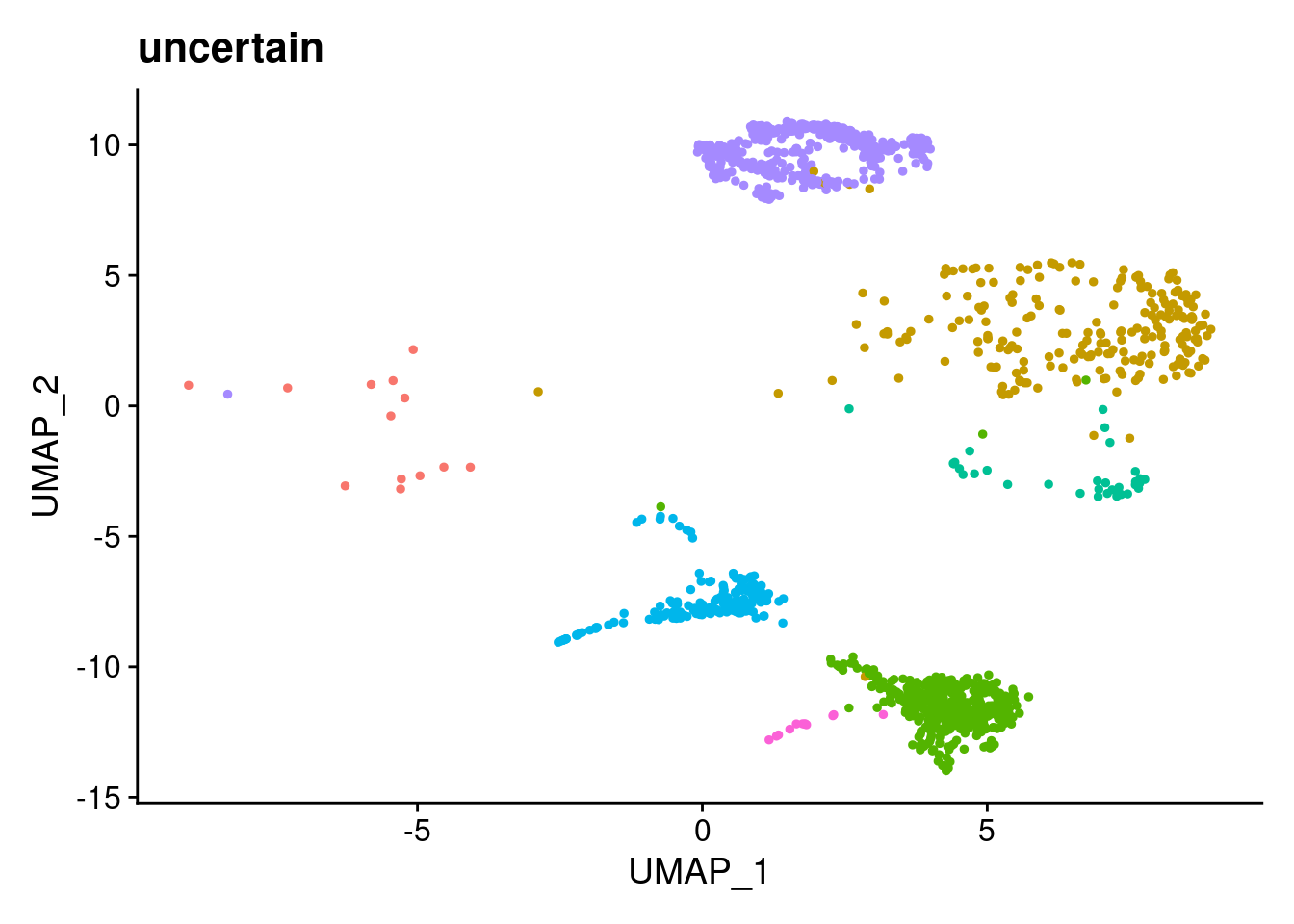
[[5]]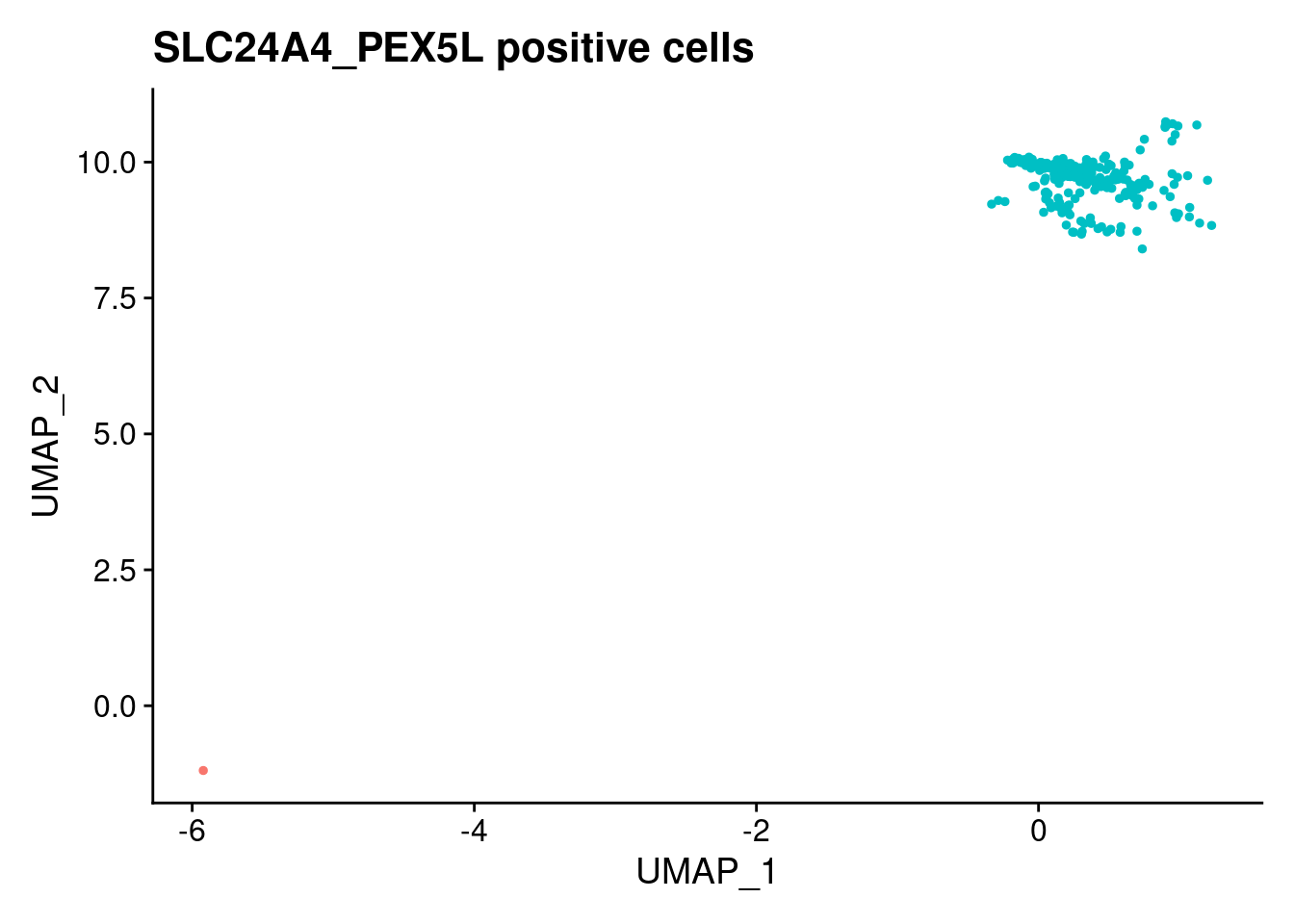
[[6]]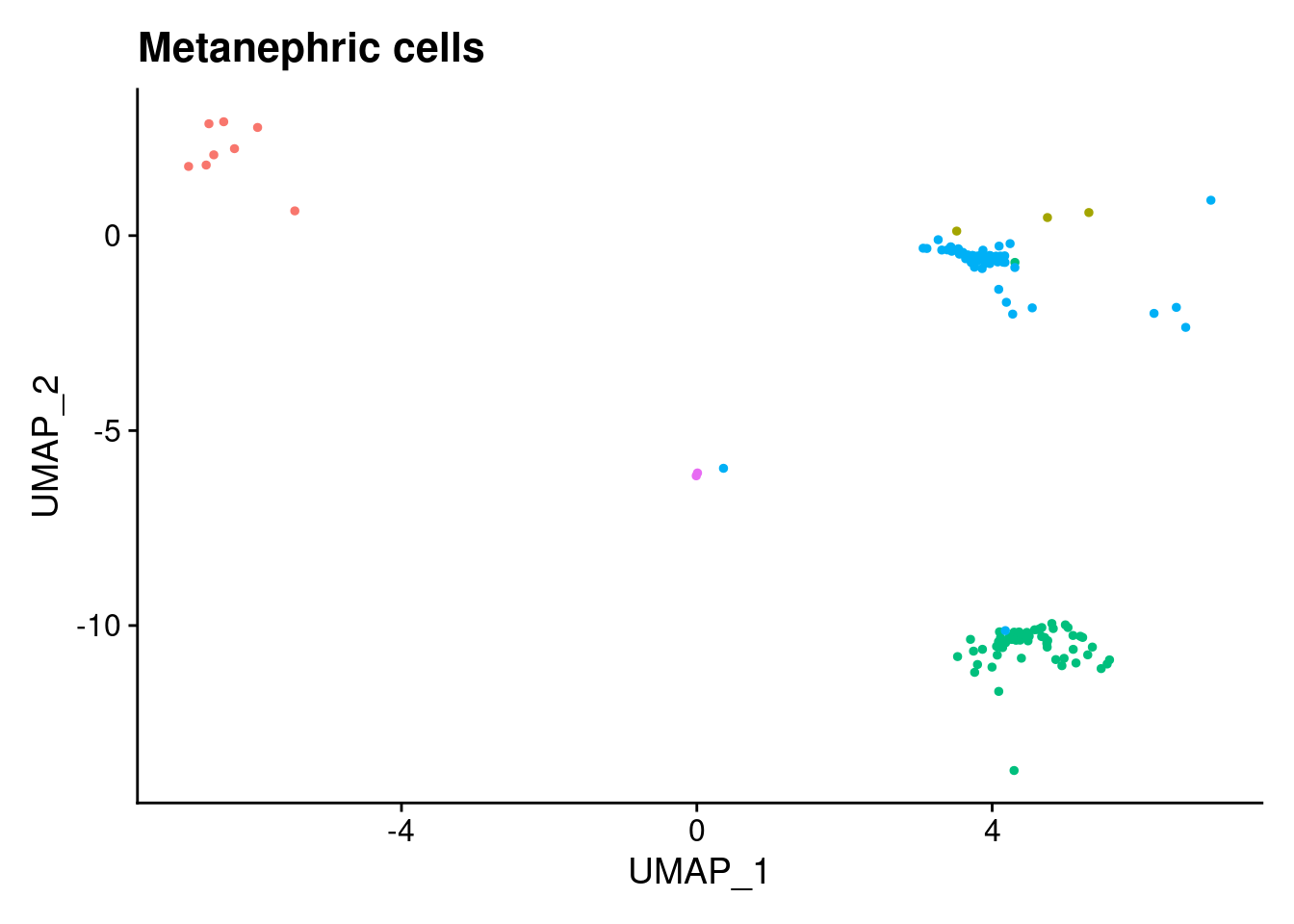
[[7]]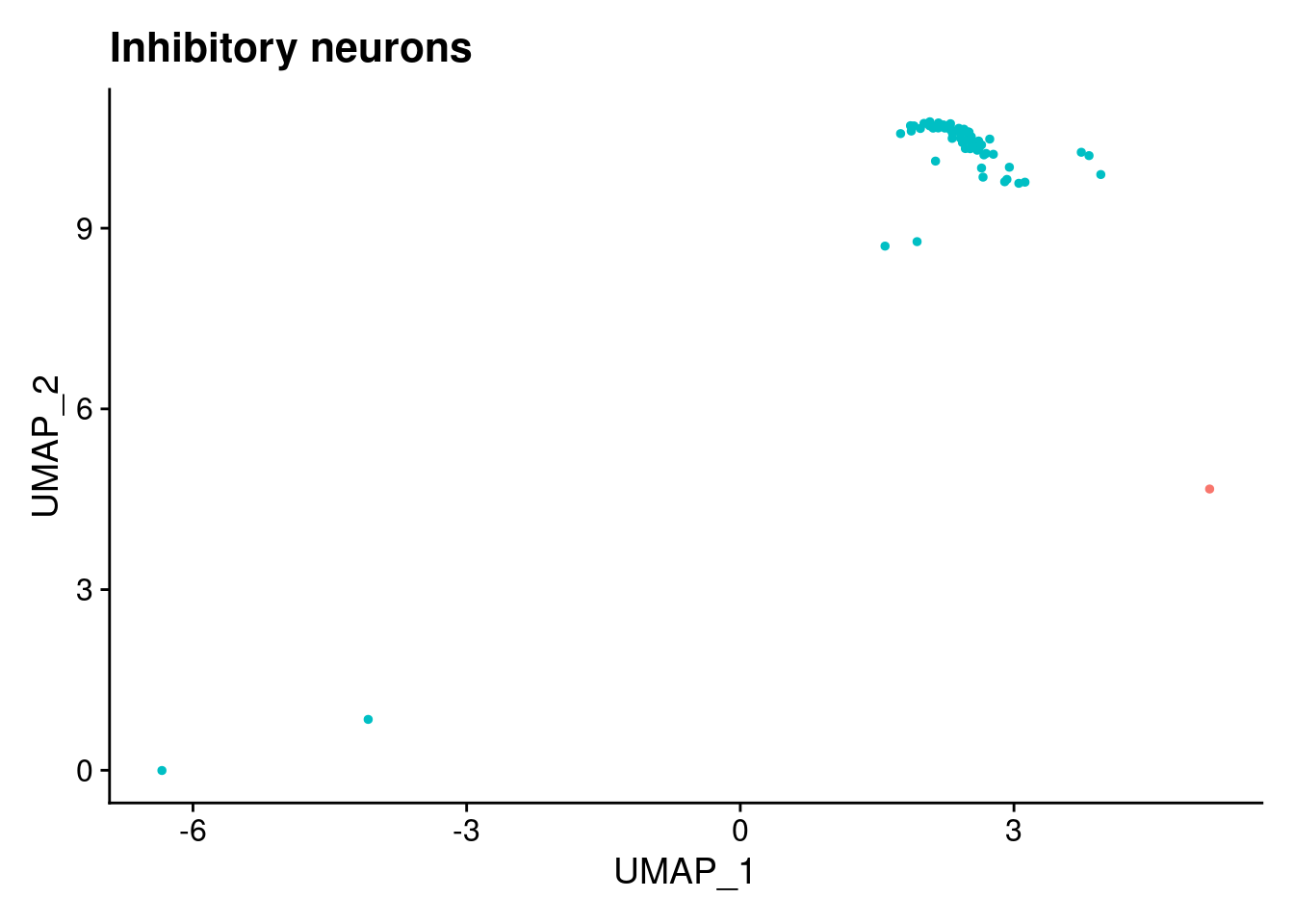
[[8]]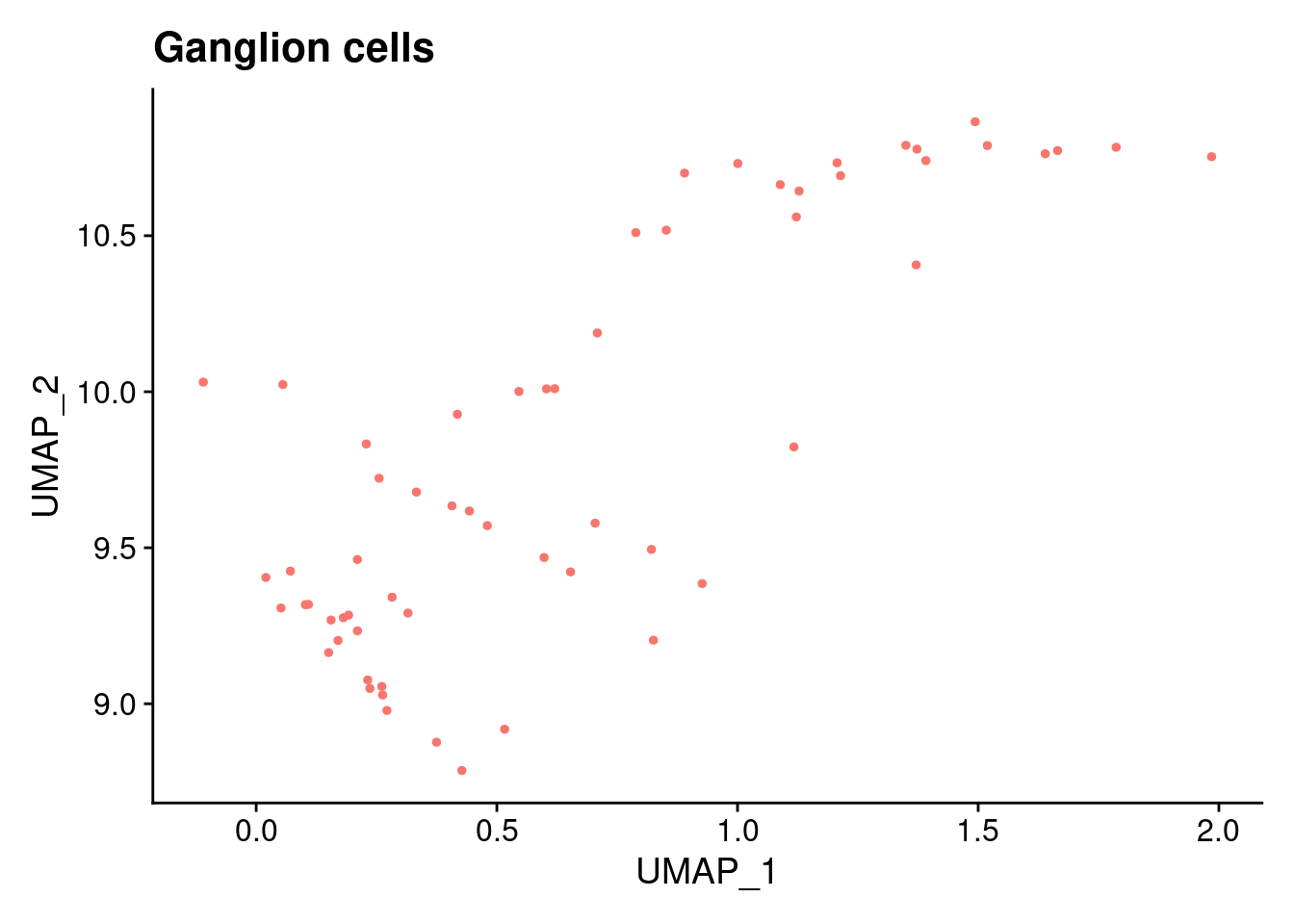
[[9]]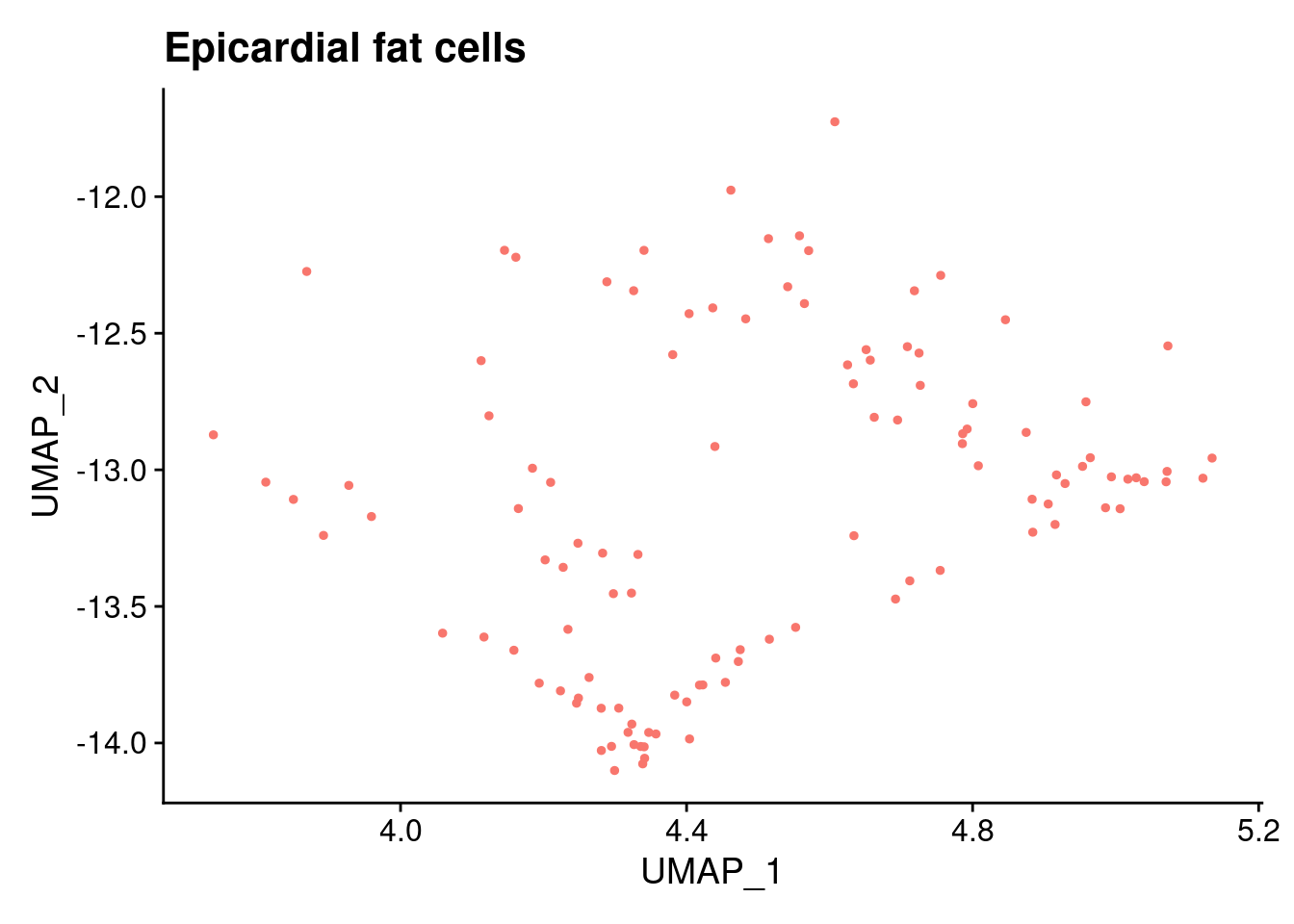
[[10]]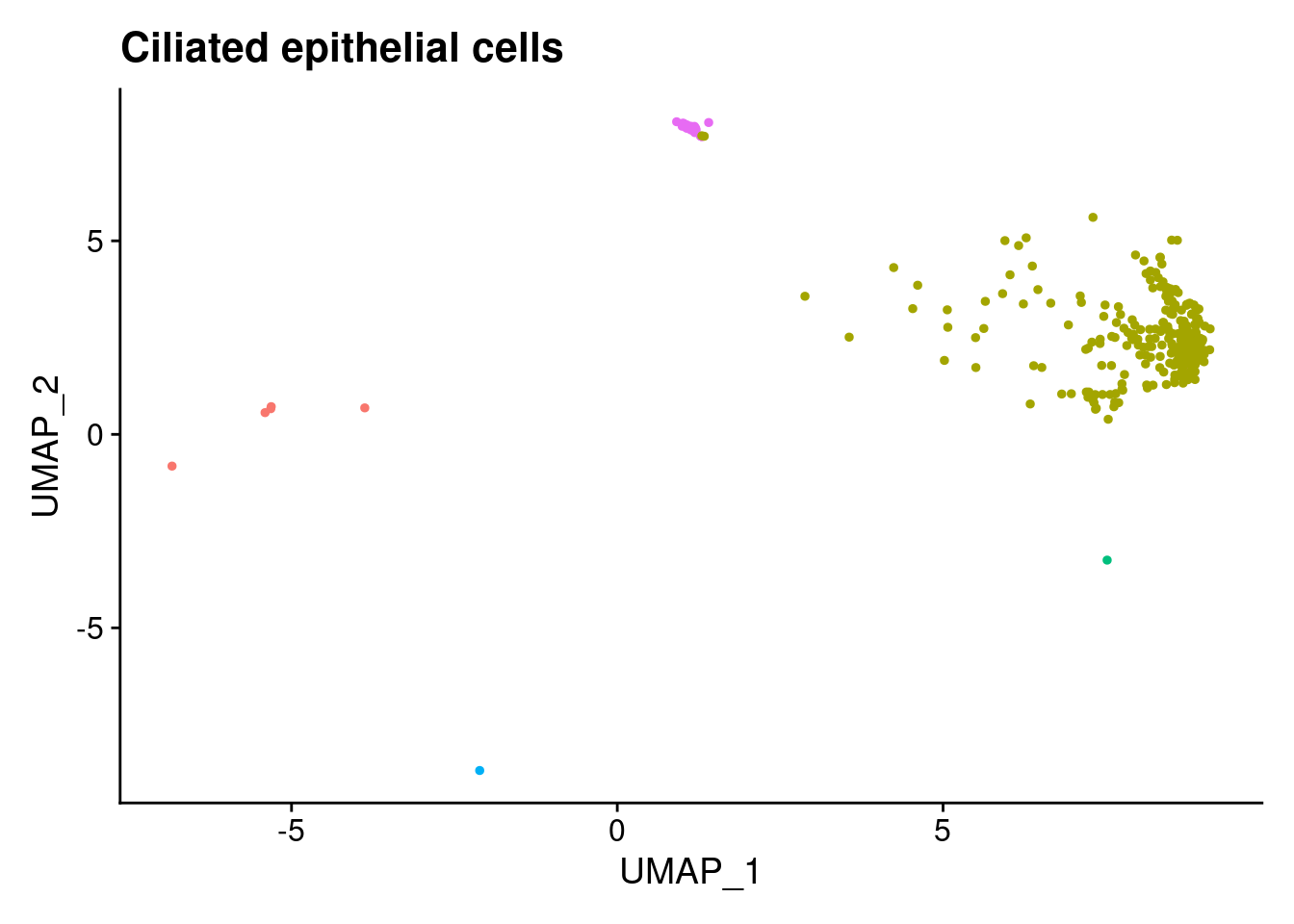
[[11]]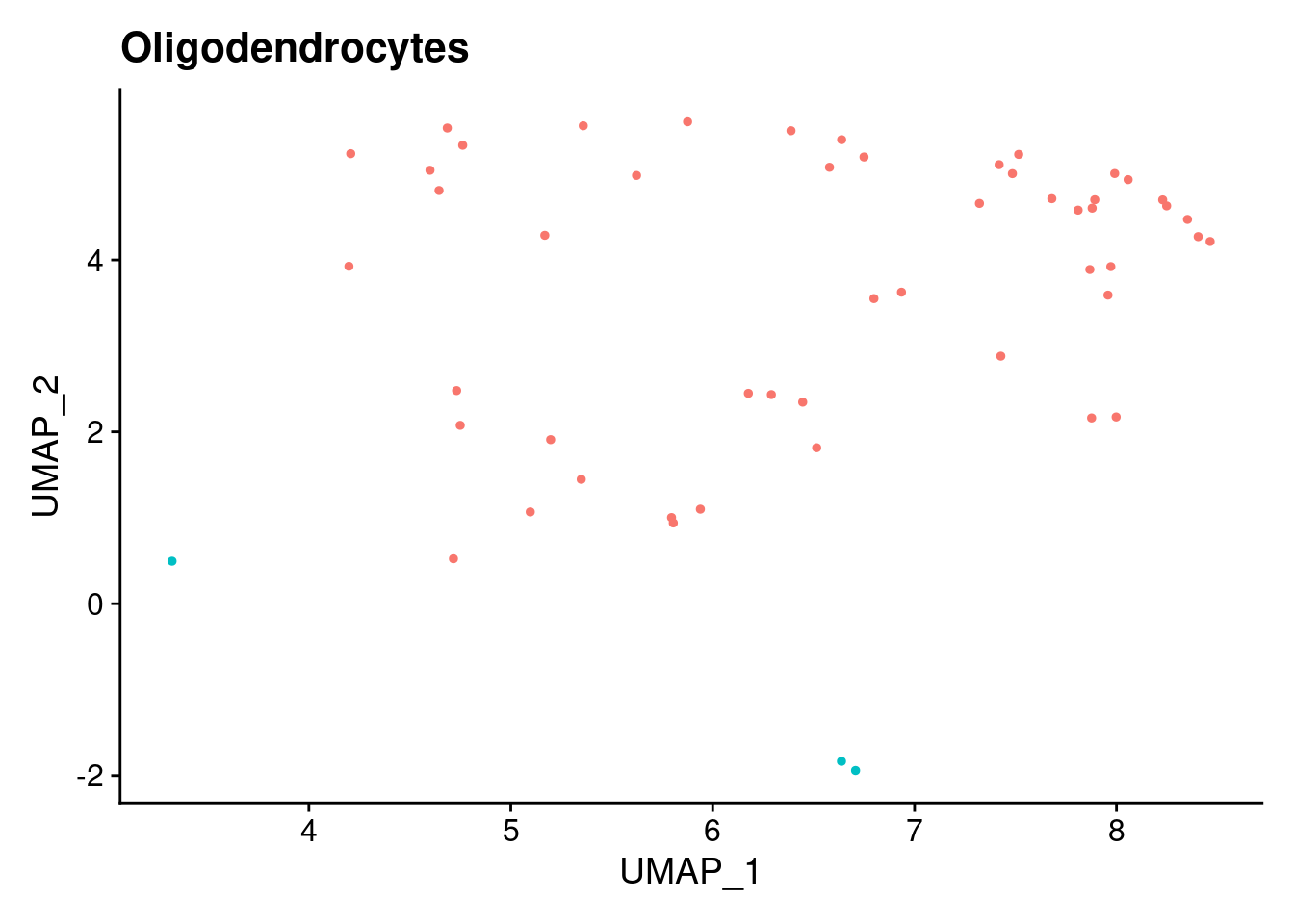
[[12]]
[[13]]
[[14]]
[[15]]
[[16]]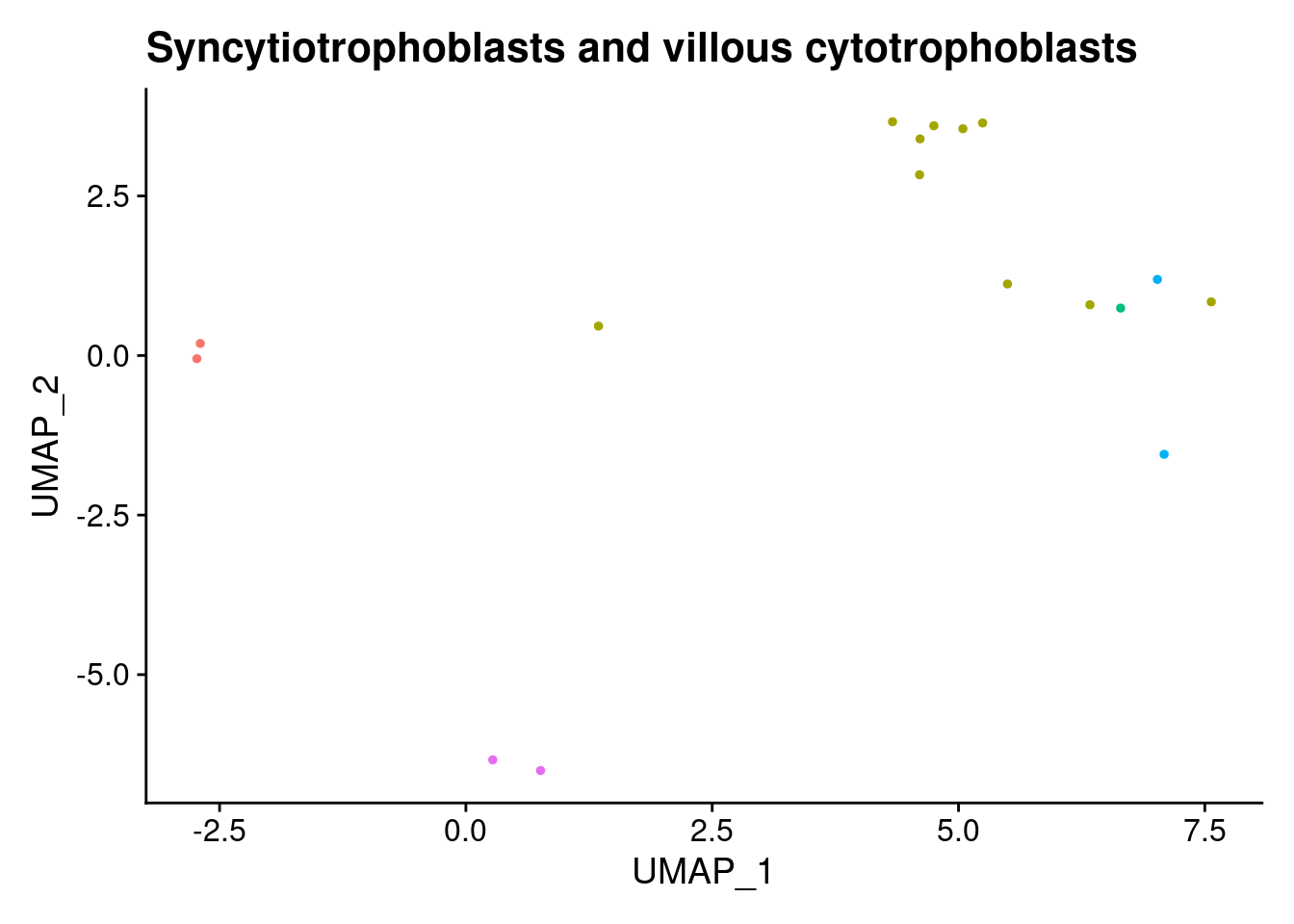
[[17]]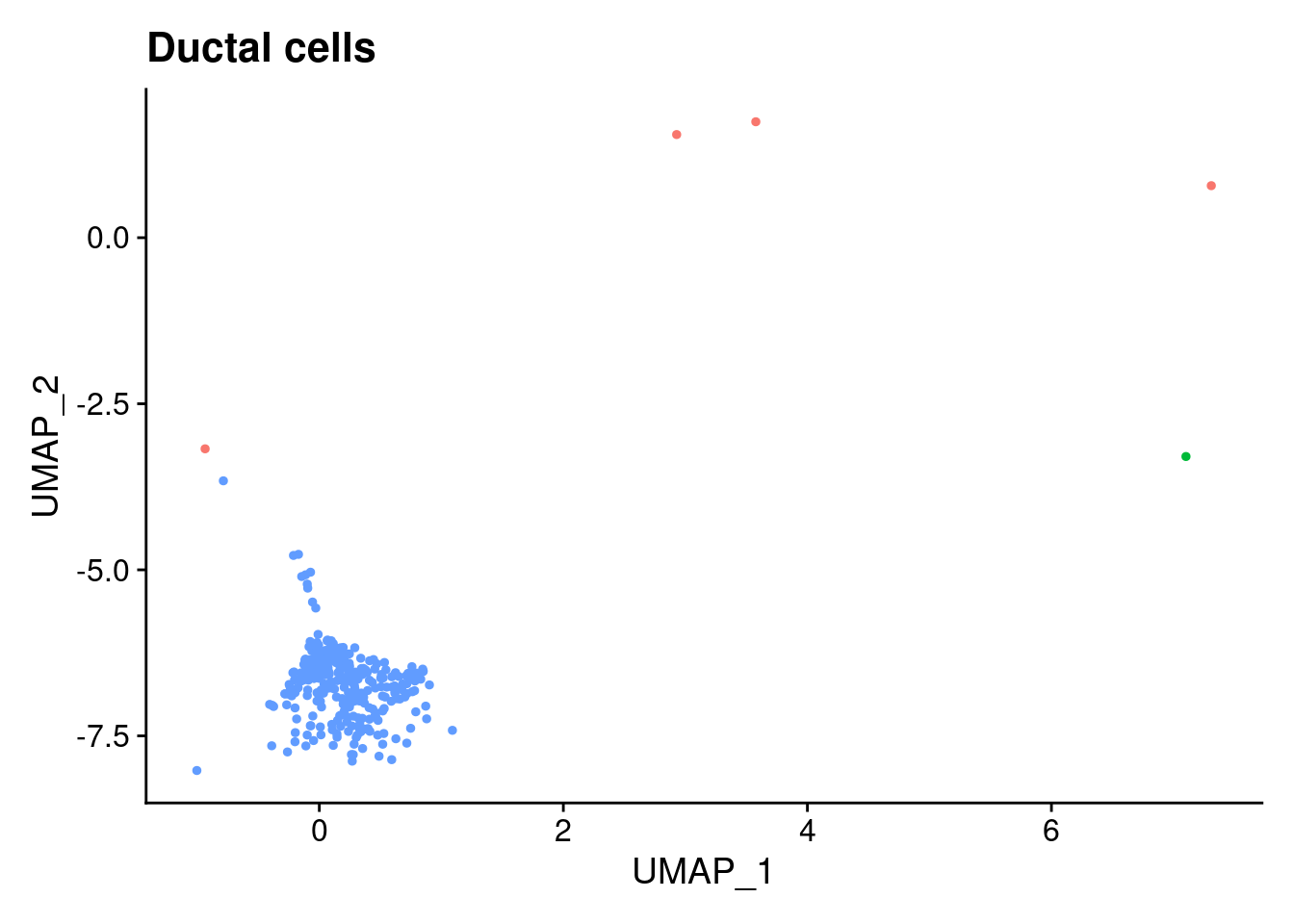
[[18]]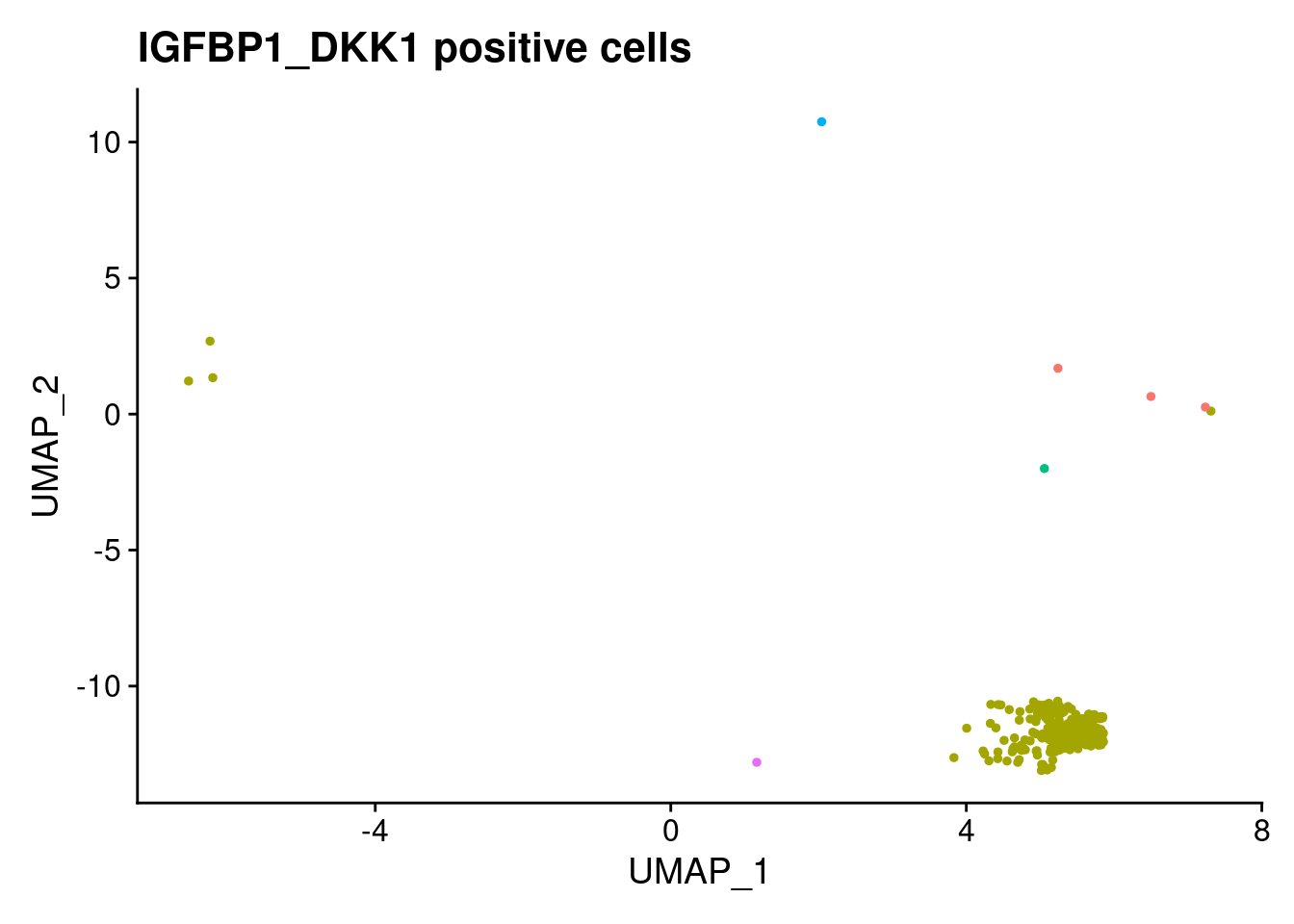
[[19]]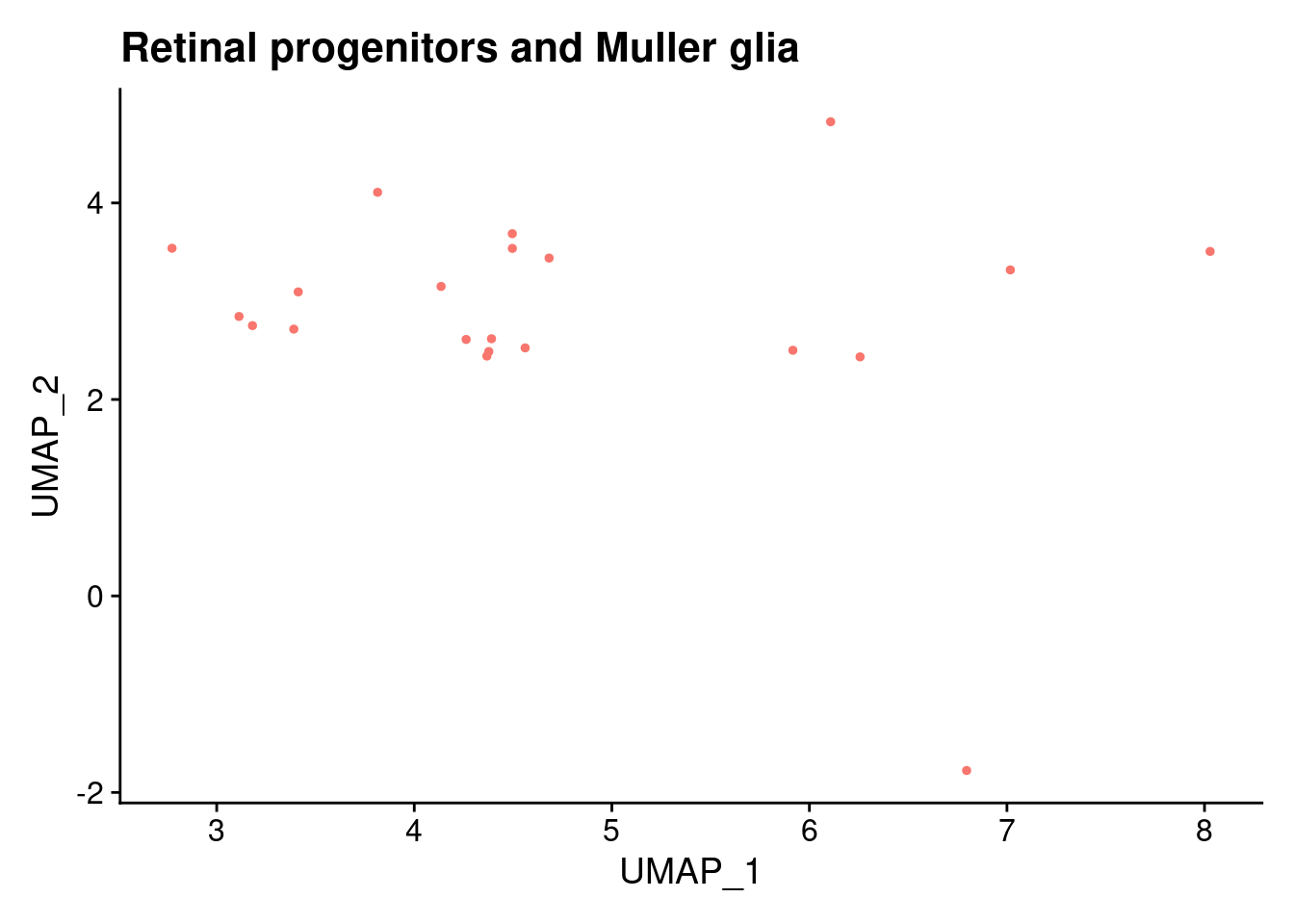
[[20]]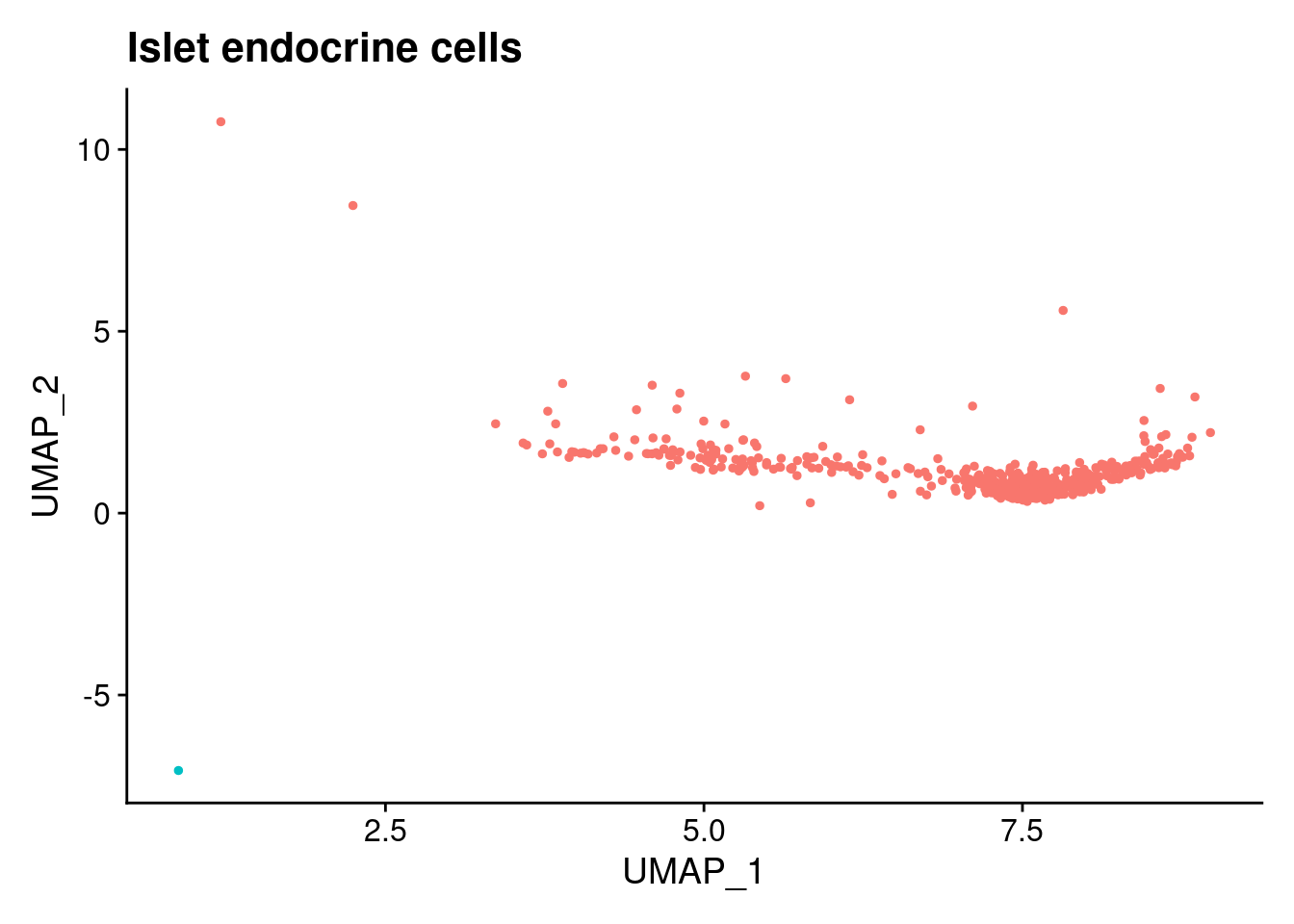
[[21]]
[[22]]
[[23]]
[[24]]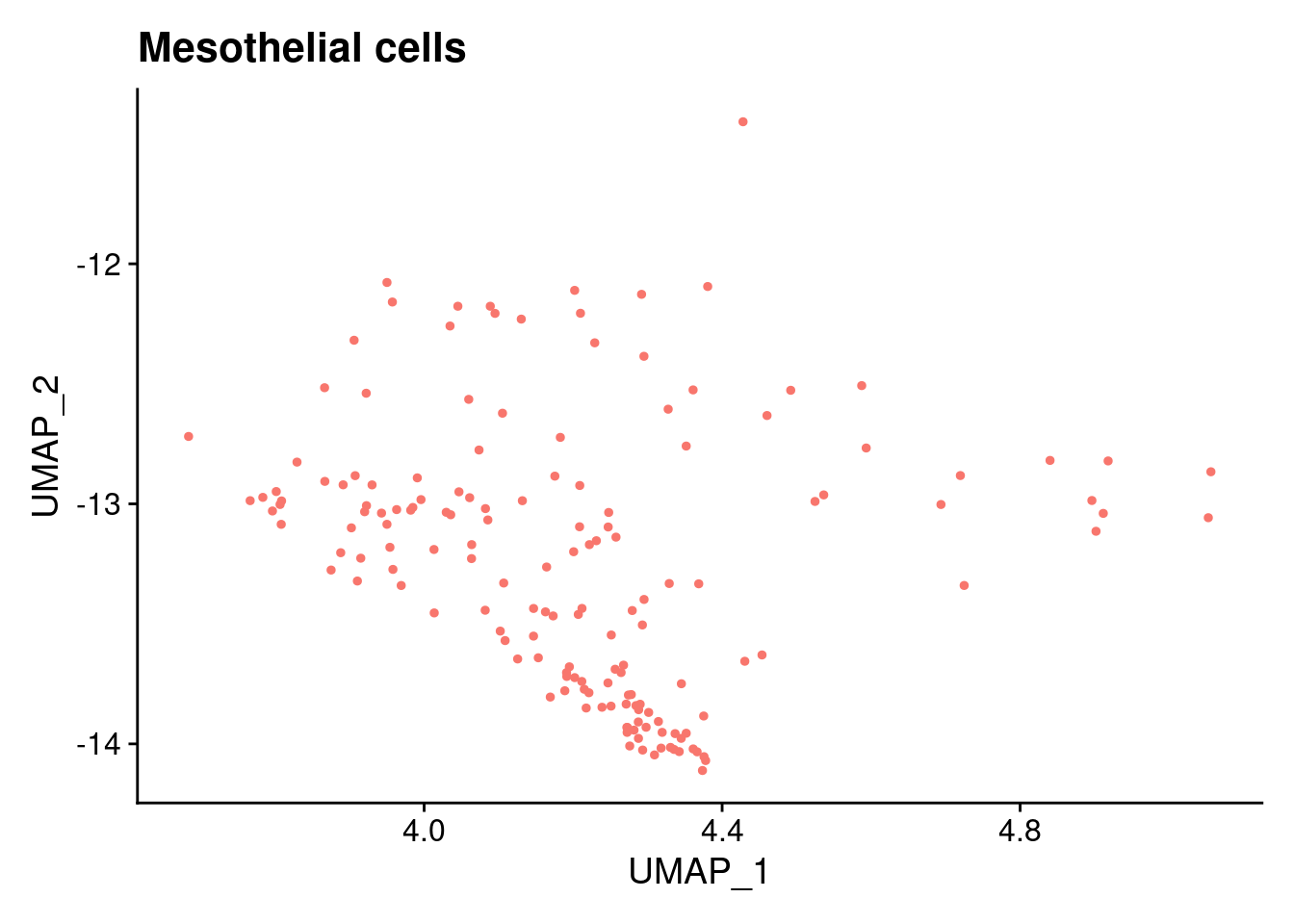
[[25]]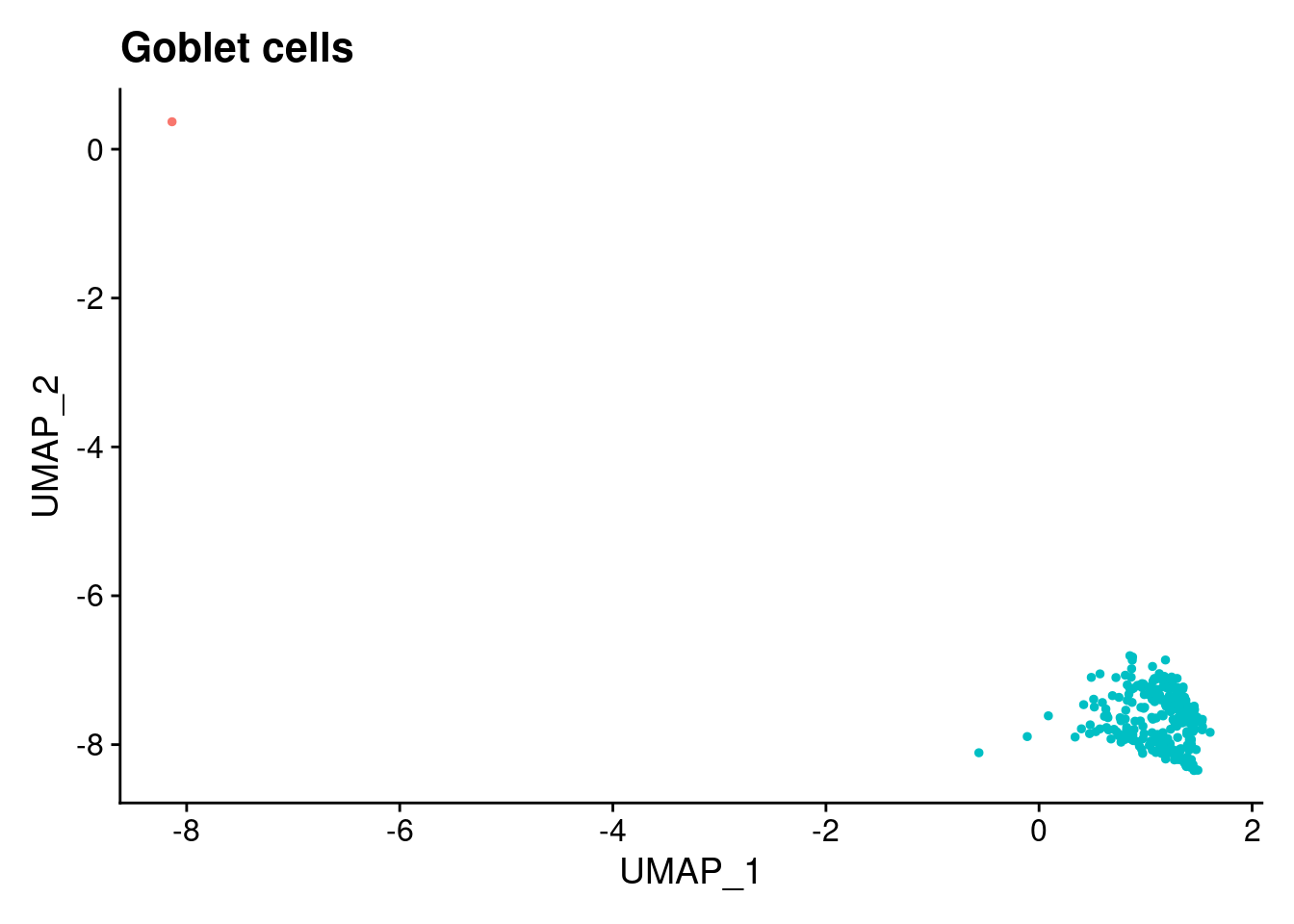
[[26]]
[[27]]
[[28]]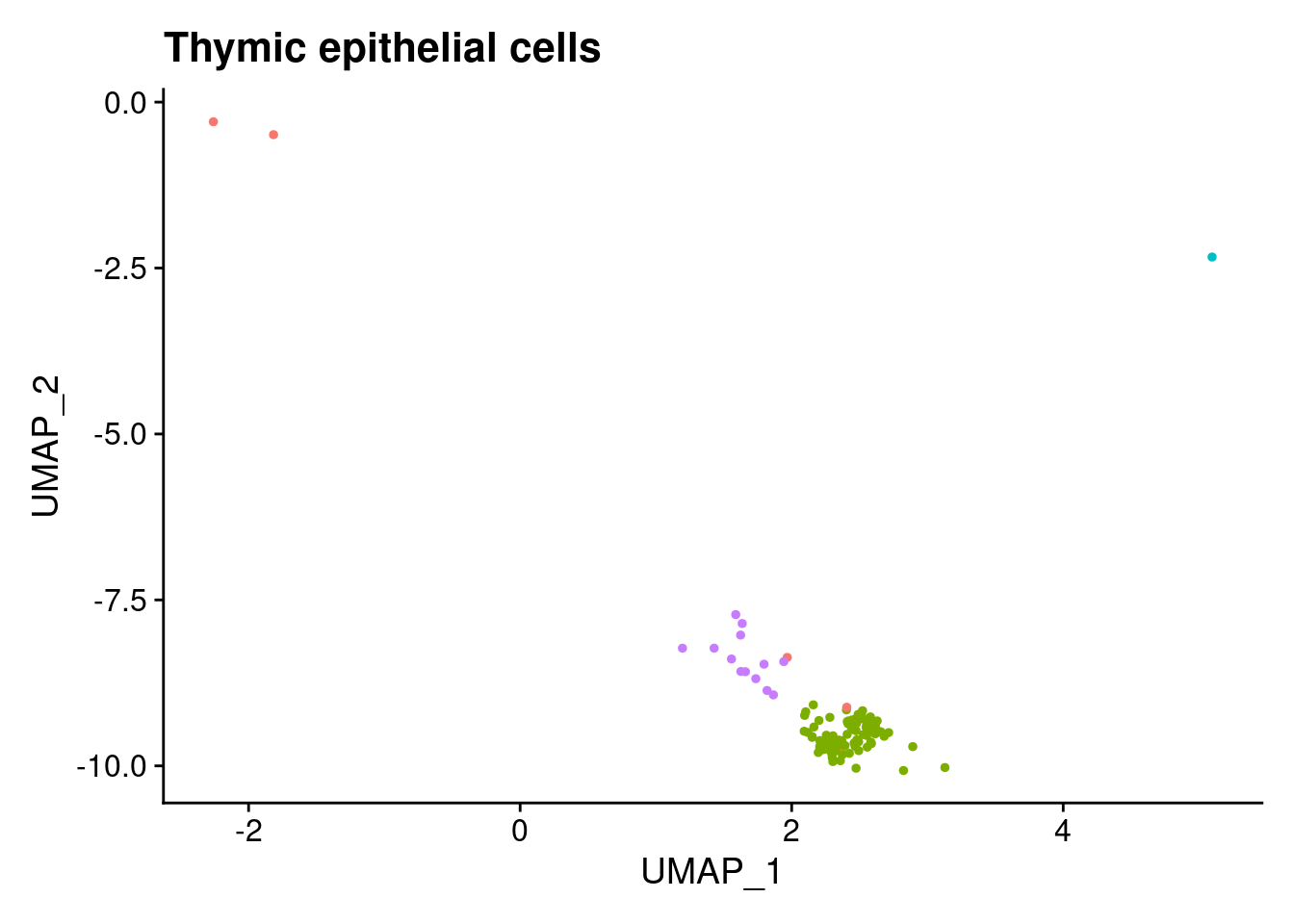
[[29]]
[[30]]
[[31]]
[[32]]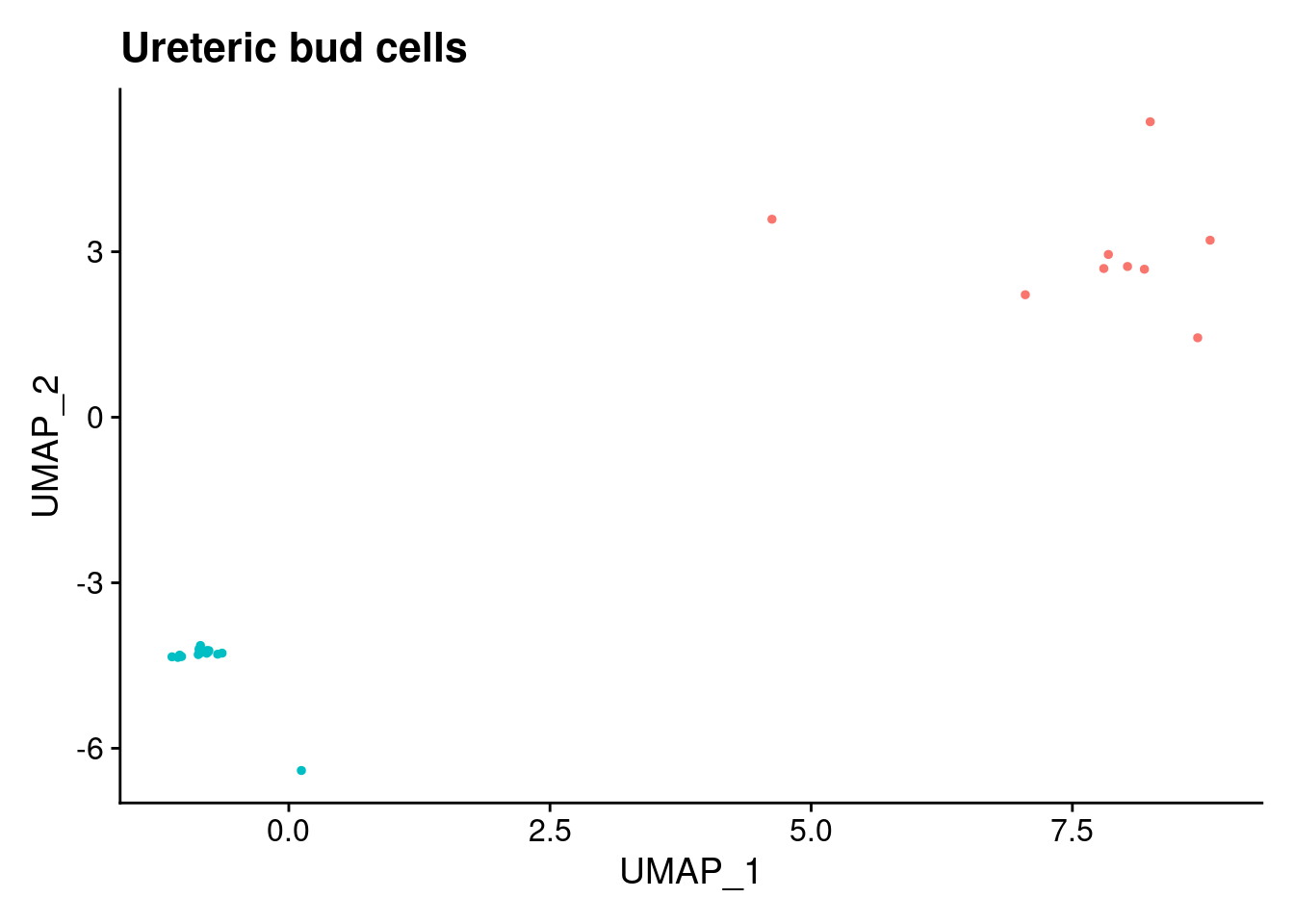
[[33]]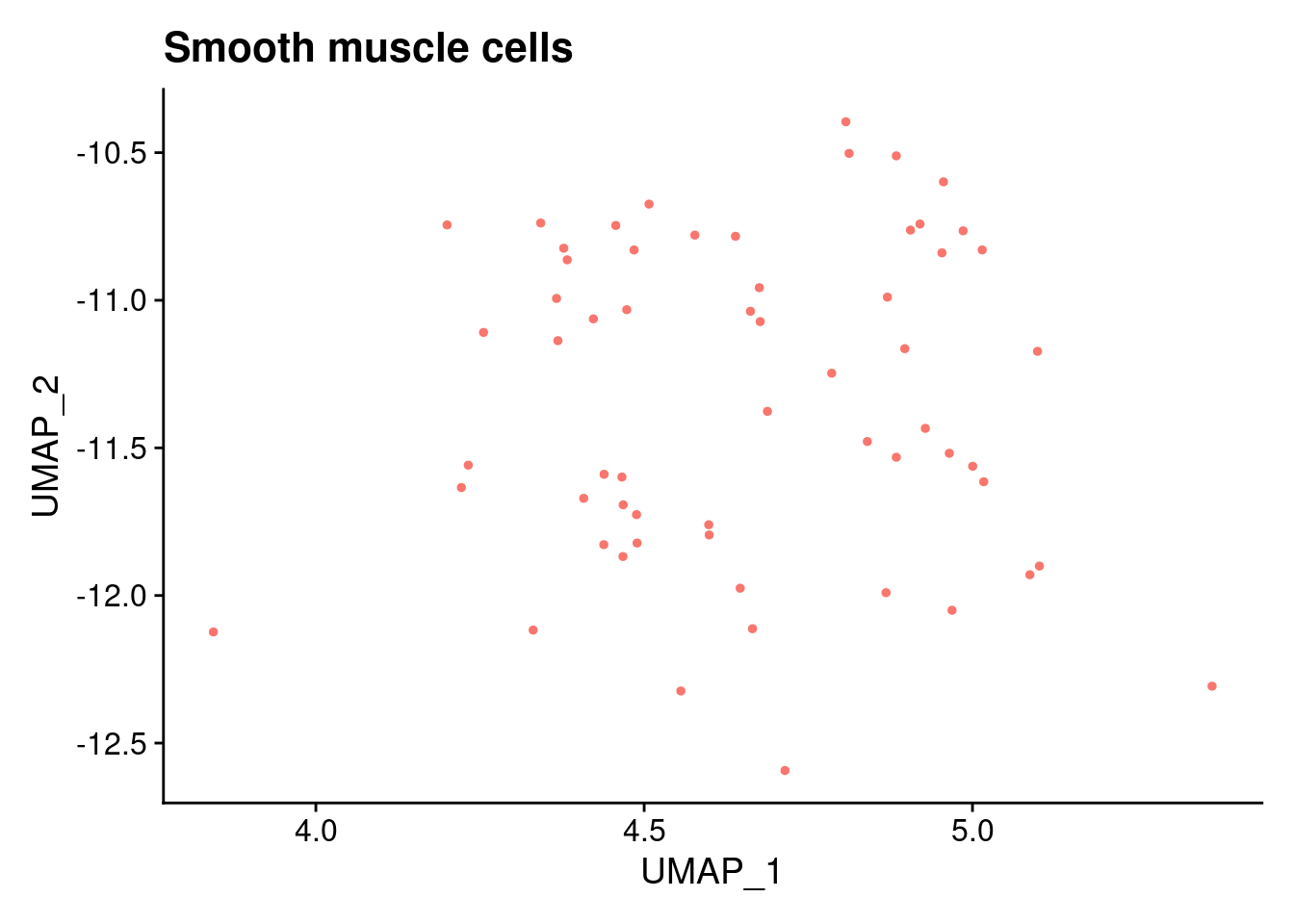
[[34]]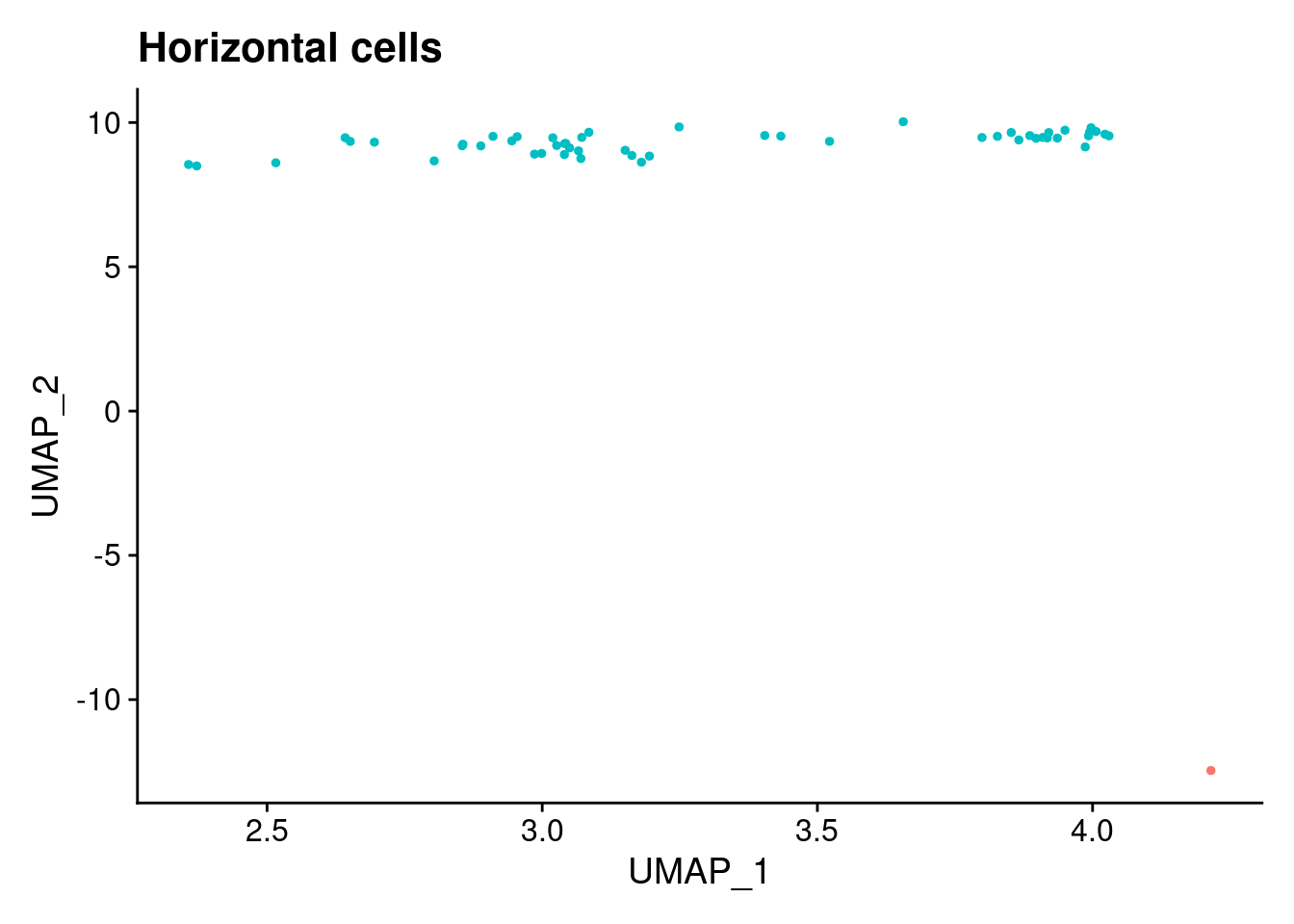
[[35]]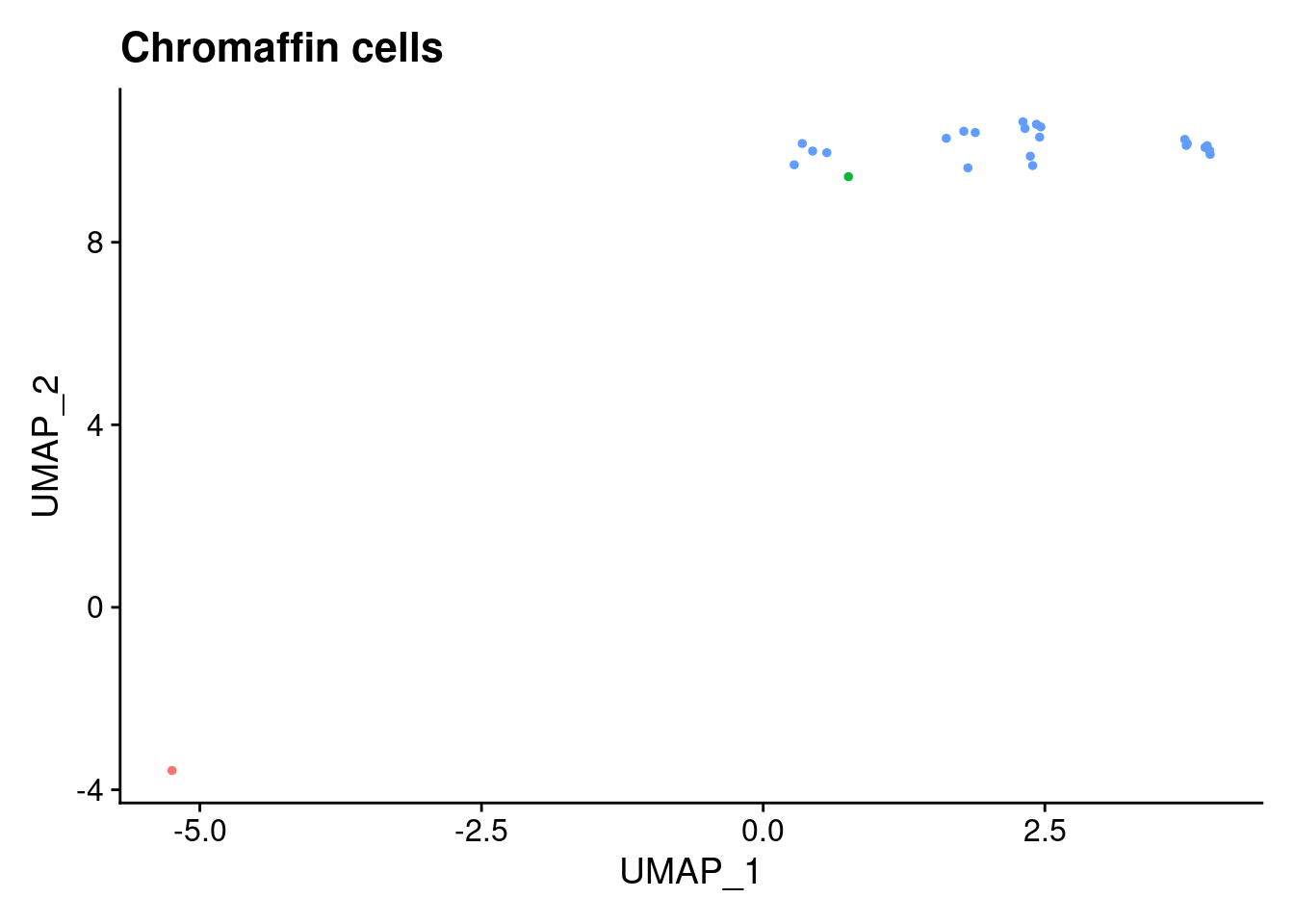
[[36]]
[[37]]
[[38]]
[[39]]
[[40]]
[[41]]
[[42]]
[[43]]
[[44]]
[[45]]
[[46]]
[[47]]
[[48]]
[[49]]
[[50]]
[[51]]
[[52]]
[[53]]
[[54]]
[[55]]
[[56]]
[[57]]
[[58]]
[[59]]
[[60]]
[[61]]
[[62]]
[[63]]
[[64]]
#Add Group Metadata
Group<- factor(paste(merged@meta.data$RefAnn, merged@meta.data$Batch, merged@meta.data$individual, sep="."))
merged<- AddMetaData(merged, Group, col.name = "Group")subset to: cells from groups that have at least 10 cells total
Ngroup<- table(merged@meta.data$Group)
Ngroup.keep<- Ngroup[Ngroup>=10]
merged<- subset(merged, subset= Group %in% names(Ngroup.keep))submerged<- as.SingleCellExperiment(merged, assay="RNA")sumex<- sumCountsAcrossCells(submerged, ids=submerged@colData$Group)
sumex<- sumex[,colSums(sumex)>0]Group<- colnames(sumex)
cluster<- as.vector(substr(Group, 1, regexpr("*.B", Group)-1))
batch<- substr(Group, regexpr("Batch", Group),regexpr("Batch", Group)+5)
ind<- substr(Group, regexpr("NA", Group),regexpr("NA", Group)+6)
samps<- cbind(cluster,batch,ind,Group)dge<- DGEList(sumex, samples=samps, remove.zeros = T)Removing 1809 rows with all zero countsremove uncertain
dge<-dge[,dge$samples$cluster != "uncertain"]keep cell types that have at least 4 groups (at least 2 individuals, at least 2 replicates)
ct.keep<- table(dge$samples$cluster)
ct.keep<- names(ct.keep[ct.keep>=4])
dge<- dge[,dge$samples$cluster %in% ct.keep]23 cell types left after this cell/celltype filtering
#filtering genes
genes.keep<- filterByExpr(dge, group= Group)
dge<- dge[genes.keep,,keep.lib.sizes=F]dge<- calcNormFactors(dge, method="TMM")dge$samples$cluster<- as.factor(as.character(dge$samples$cluster))#not including batch and individual in model because I do not have balances representation across cell types
design<- model.matrix(~0+ dge$samples$cluster)v<- voom(dge, design, plot=T)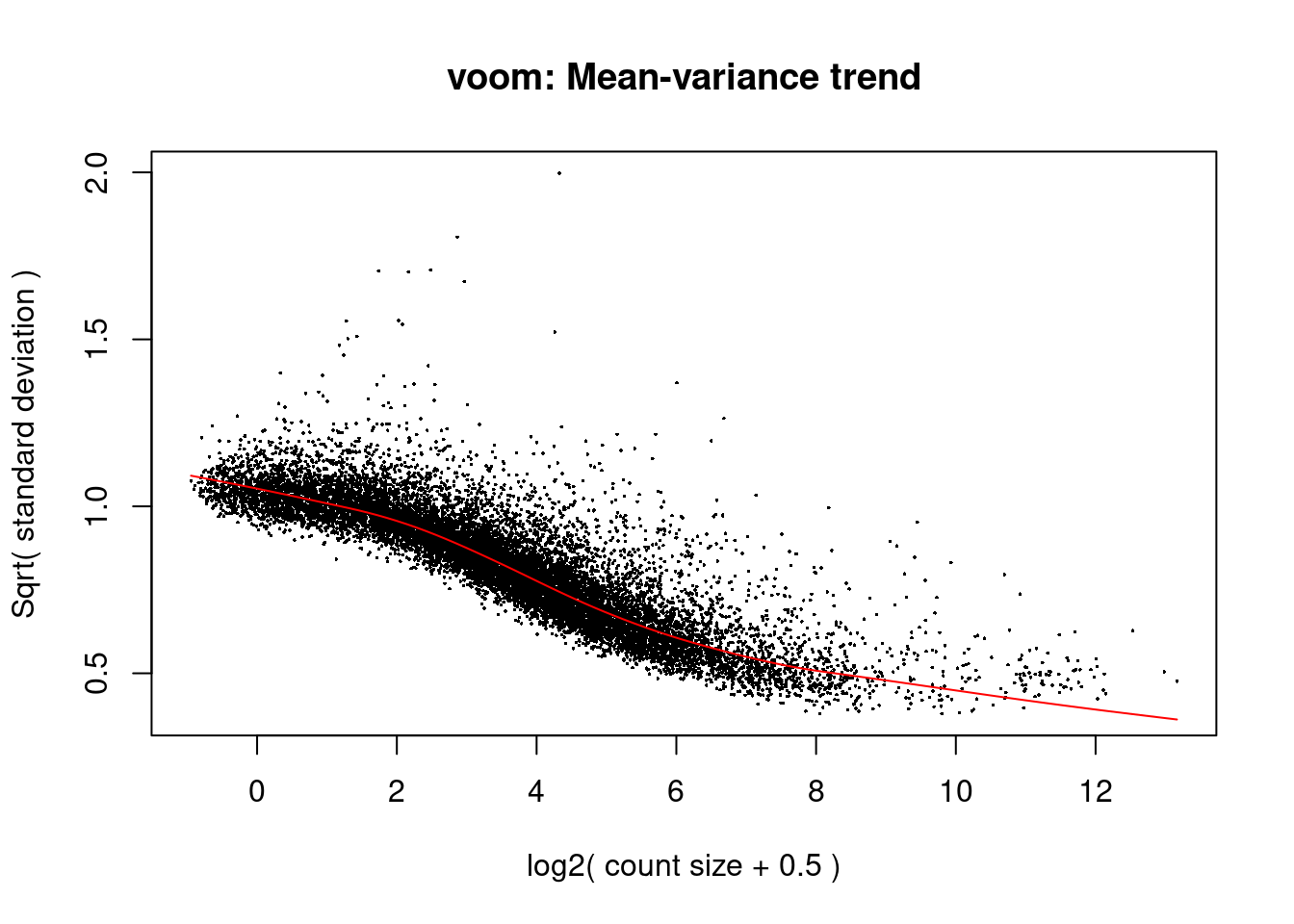
fit<- lmFit(v,design)nclust<- length(unique(dge$samples$cluster))
contrasts<- NULL
for (i in 1:nclust){
c<- c(rep(-1,nclust))
c[i]<- nclust-1
contrasts<- cbind(contrasts, c)
}contrasts c c c c c c c c c c c c c c c c c c c c c c c
[1,] 22 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1
[2,] -1 22 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1
[3,] -1 -1 22 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1
[4,] -1 -1 -1 22 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1
[5,] -1 -1 -1 -1 22 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1
[6,] -1 -1 -1 -1 -1 22 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1
[7,] -1 -1 -1 -1 -1 -1 22 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1
[8,] -1 -1 -1 -1 -1 -1 -1 22 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1
[9,] -1 -1 -1 -1 -1 -1 -1 -1 22 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1
[10,] -1 -1 -1 -1 -1 -1 -1 -1 -1 22 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1
[11,] -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 22 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1
[12,] -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 22 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1
[13,] -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 22 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1
[14,] -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 22 -1 -1 -1 -1 -1 -1 -1 -1 -1
[15,] -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 22 -1 -1 -1 -1 -1 -1 -1 -1
[16,] -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 22 -1 -1 -1 -1 -1 -1 -1
[17,] -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 22 -1 -1 -1 -1 -1 -1
[18,] -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 22 -1 -1 -1 -1 -1
[19,] -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 22 -1 -1 -1 -1
[20,] -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 22 -1 -1 -1
[21,] -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 22 -1 -1
[22,] -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 22 -1
[23,] -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 22fit<- contrasts.fit(fit, contrasts=contrasts)efit<- eBayes(fit)plotSA(efit)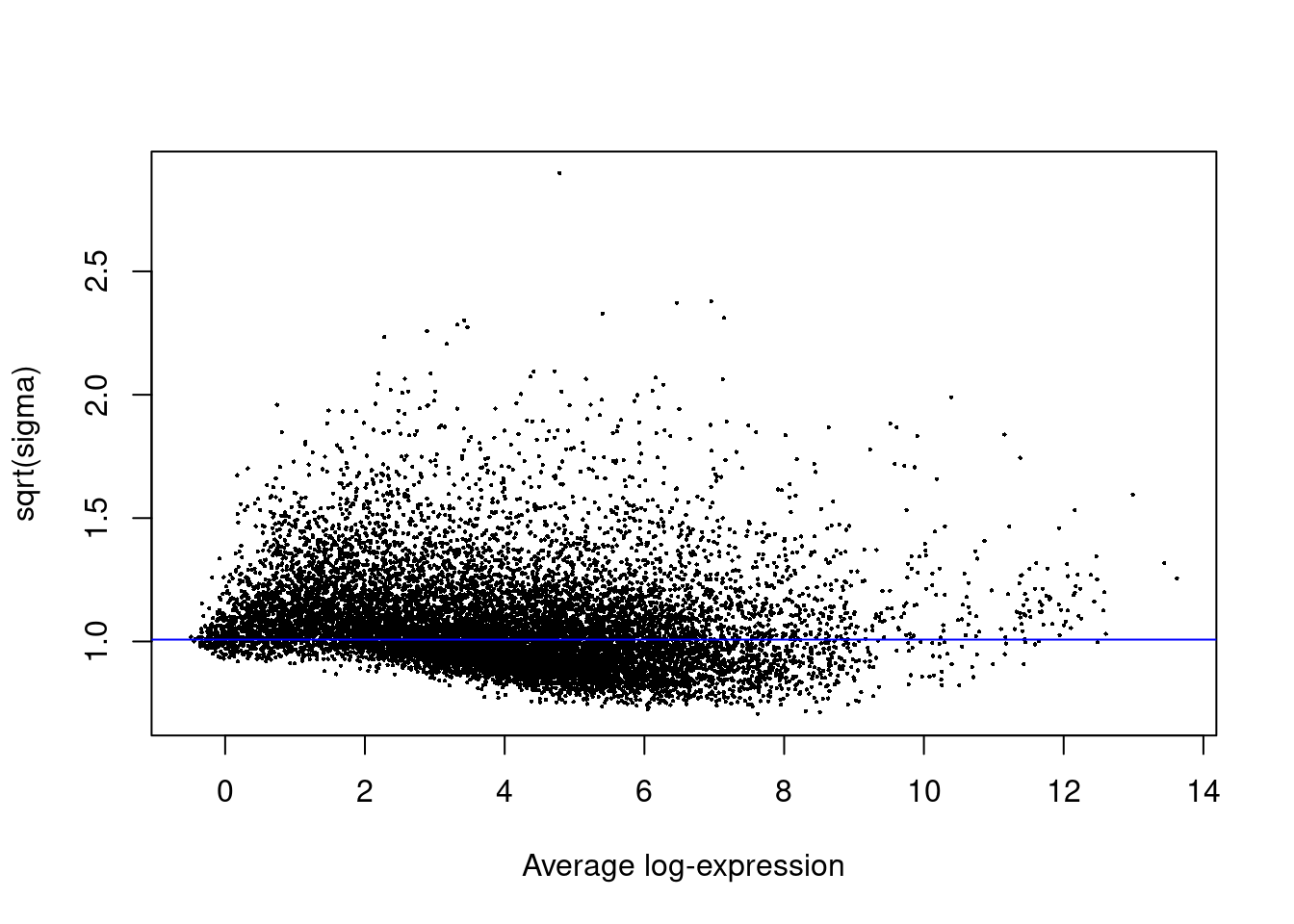
summary(decideTests(efit)) c c c c c c c c c c c c
Down 271 1438 658 620 559 1067 1723 3322 479 1891 920 183
NotSig 11839 9768 11061 10995 11140 10216 9226 6595 11280 8806 10485 11993
Up 971 1875 1362 1466 1382 1798 2132 3164 1322 2384 1676 905
c c c c c c c c c c c
Down 376 1190 91 643 2549 979 792 364 144 560 5167
NotSig 11533 9989 12383 11051 7843 10224 10696 11574 12148 11091 3273
Up 1172 1902 607 1387 2689 1878 1593 1143 789 1430 4641levels(dge$samples$cluster) [1] "Amacrine cells" "Cardiomyocytes"
[3] "Ciliated epithelial cells" "Ductal cells"
[5] "Epicardial fat cells" "Goblet cells"
[7] "Granule neurons" "Hepatoblasts"
[9] "IG" "Intestinal epithelial cells"
[11] "Islet endocrine cells" "MUC13_D"
[13] "Mesangial cells" "Mesothelial cells"
[15] "Metanephric cells" "Retinal pigment cells"
[17] "SLC24A4_PEX5L positive cells" "Squamous epithelial cells"
[19] "Stellate cells" "Stromal cells"
[21] "Thymic epithelial cells" "Visceral neurons"
[23] "scHCL.hESC" For example, lets see the top genes DE by Cardiomyocytes (coef 2)
topTable(efit, coef=2, sort.by="P", n=50) logFC AveExpr t P.Value adj.P.Val B
MYL7 130.66430 3.0976950 30.21845 1.847703e-54 2.416980e-50 46.30499
BMP4 100.17794 4.6916506 26.04480 2.482904e-48 1.244905e-44 47.33820
MYL4 128.60340 3.2151048 26.00579 2.855068e-48 1.244905e-44 43.38726
CYB5D1 83.34643 3.8300429 25.22105 4.896782e-47 1.601370e-43 38.70436
APOBEC3G 90.85741 2.0962458 23.72382 1.325661e-44 3.240680e-41 27.97143
DOK4 71.25664 4.8213956 23.69388 1.486437e-44 3.240680e-41 41.78124
CFC1 100.08962 1.1041893 22.03796 9.765033e-42 1.824806e-38 23.39502
HS3ST3A1 86.60612 2.1116755 21.92962 1.509262e-41 2.467832e-38 26.51592
GLIPR2 73.08017 3.9350171 21.68732 4.016454e-41 5.837692e-38 35.41663
BAG3 58.11844 4.2447476 21.03112 5.891112e-40 7.706163e-37 33.46104
FGF19 107.72012 2.2553657 20.82809 1.366447e-39 1.624954e-36 29.36165
BMPER 96.84503 2.3372969 20.16516 2.207314e-38 2.406156e-35 28.62389
TNNT2 92.88780 2.9350416 20.11749 2.701837e-38 2.718672e-35 30.65986
COBLL1 66.26504 3.8078148 18.55705 2.360551e-35 2.205597e-32 28.89878
DNAH2 87.33649 1.5861408 18.34031 6.195401e-35 5.402803e-32 20.95438
KRT19 65.97059 8.6797897 18.26668 8.610261e-35 7.039426e-32 49.52404
PLPPR5 84.77348 1.3670778 18.18679 1.231576e-34 9.476617e-32 18.83396
TMOD1 83.64226 1.9293714 18.15105 1.445832e-34 1.050718e-31 21.66704
PSKH2 102.71186 0.5888992 18.12316 1.638795e-34 1.078437e-31 18.74104
APLNR 121.89089 2.3444355 18.12179 1.648860e-34 1.078437e-31 29.79227
TMEM185A 53.09470 3.9425363 18.02878 2.505759e-34 1.506125e-31 27.13528
TNNI1 121.51287 3.9575031 18.02637 2.533044e-34 1.506125e-31 35.69392
RGS13 144.64986 2.6808708 17.99972 2.856335e-34 1.568971e-31 32.12976
HTRA1 80.66431 3.5987240 17.99800 2.878626e-34 1.568971e-31 29.75697
PRDM6 81.90209 1.8387960 17.78189 7.650104e-34 4.002840e-31 20.50841
LHFPL2 51.42098 3.8578678 17.43567 3.707501e-33 1.865301e-30 25.51257
ALPK2 86.60294 2.0494678 17.39412 4.485349e-33 2.173068e-30 22.24025
HAND1 134.92284 4.6368572 17.30063 6.890111e-33 3.218912e-30 37.49295
LYN 58.36667 3.5424557 17.05154 2.174038e-32 9.806410e-30 24.28785
SVEP1 71.29496 2.2052858 16.82727 6.159146e-32 2.685593e-29 19.22263
BAMBI 74.02825 6.5277939 16.79757 7.073195e-32 2.984660e-29 39.16688
SLC9A3R1 45.81782 6.6814823 16.76133 8.375453e-32 3.423728e-29 36.82614
HAS2 87.30497 5.5998335 16.66846 1.292503e-31 5.102493e-29 36.31877
ACTC1 112.70701 5.6095105 16.66295 1.326235e-31 5.102493e-29 40.55194
FABP5 58.77926 7.8679014 16.54164 2.341450e-31 8.751003e-29 44.09118
S100A11 45.05607 10.0171268 16.52148 2.573913e-31 9.352597e-29 48.96027
SHISAL2B 73.73926 2.6002339 16.10512 1.838141e-30 6.498575e-28 20.38247
TCF21 85.51477 1.8131395 15.90283 4.814762e-30 1.657419e-27 18.99167
FAM89A 76.32724 5.8430913 15.81296 7.397133e-30 2.481074e-27 36.12206
GYPB 109.95160 0.4211041 15.66734 1.486511e-29 4.861262e-27 16.12666
PPFIBP1 40.37845 5.1133169 15.50954 3.175936e-29 1.013279e-26 27.98998
TUBB6 57.58937 5.2111883 15.47312 3.785710e-29 1.179069e-26 29.67380
ADAM19 68.91500 3.7111987 15.43315 4.591396e-29 1.396745e-26 24.78315
ADGRA2 71.74334 2.0801881 15.35020 6.856677e-29 2.036366e-26 17.12242
HAPLN1 135.31468 5.1626583 15.34577 7.005309e-29 2.036366e-26 35.92405
MIXL1 109.03120 2.2806485 15.11656 2.131129e-28 6.060284e-26 22.94231
P3H2 58.24763 3.7355420 14.89919 6.156338e-28 1.713427e-25 22.47444
RGS5 89.58711 5.7892420 14.89128 6.399314e-28 1.743946e-25 33.65425
KDR 86.16713 2.9812211 14.69530 1.673609e-27 4.467853e-25 22.30438
HOXB6 110.90093 2.6245312 14.68790 1.735630e-27 4.540755e-25 24.16276We see recognizeable markers of CMs in that list-- MYL7, MYL4, TNNT2
vol<- topTable(efit, coef=2, n=nrow(fit))
labsig<- vol[rownames(vol) %in% c("MYL7", "MYL4", "TNNT2"),]
labsiggenes<- rownames(labsig)
thresh<- vol$adj.P.Val < 0.05
vol<-cbind(vol, thresh)
v<- ggplot(vol, aes(x=logFC, y= -log10(adj.P.Val))) +
geom_point(aes(colour=thresh), show.legend = FALSE) +
scale_colour_manual(values = c("TRUE" = "red", "FALSE" = "black")) +
geom_text(data=labsig, aes(label=labsiggenes))
v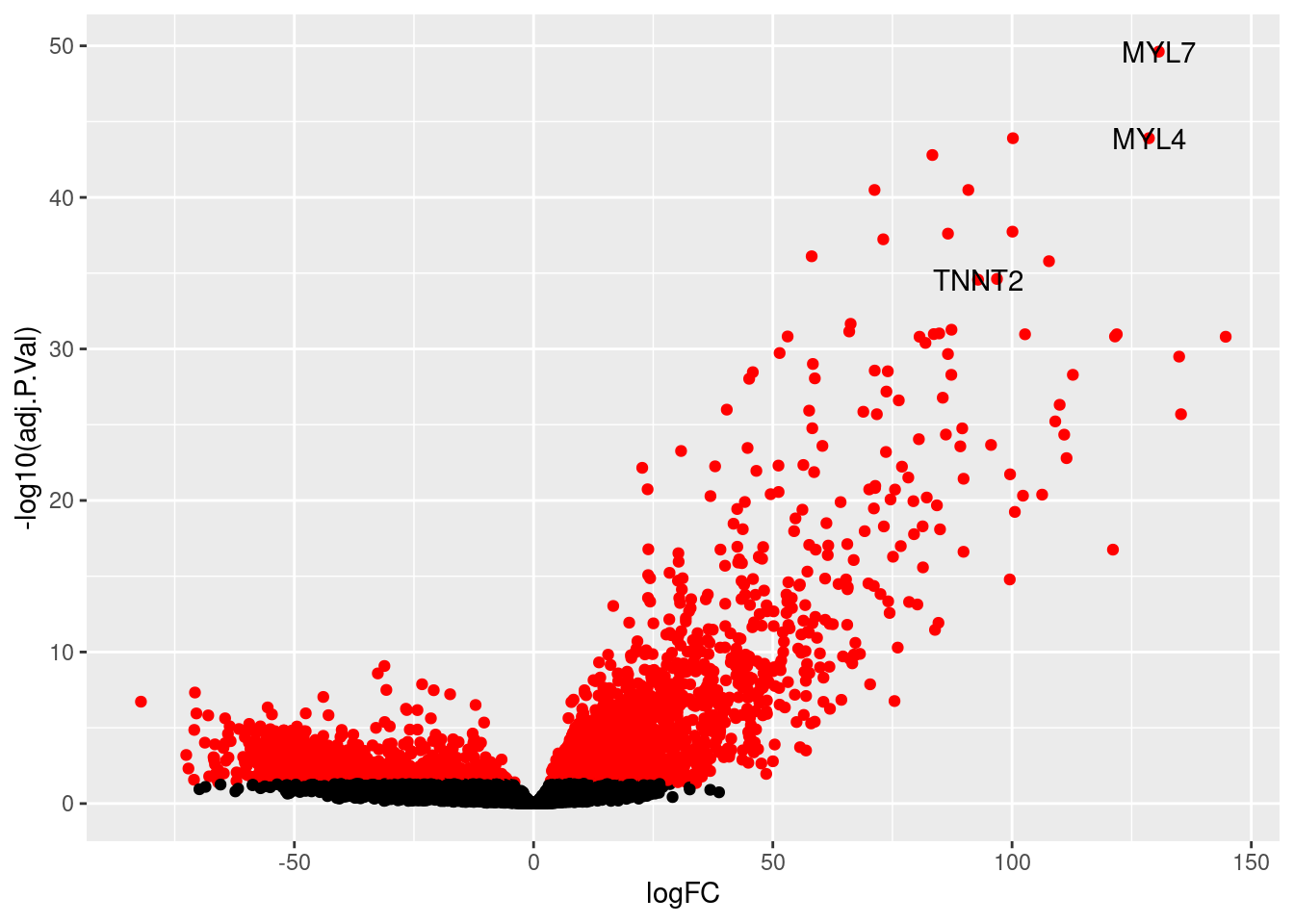
png(file= "/project2/gilad/katie/Pilot_HumanEBs/Embryoid_Body_Pilot_Workflowr/output/figs/Supp_RefAnnDE_cardiomyocyte.png", width= 6, height=6, units= "in", res= 1080)
v
dev.off()make list of top 20 most significant DE genes in each of the 23 cell types in this analysis
Top20DE<- NULL
for (i in 1:nclust){
Top20DE[[i]]<- topTable(efit, coef=i, sort.by="P", n=50)
}
names(Top20DE)<- levels(dge$samples$cluster)
Top20DE$`Amacrine cells`
logFC AveExpr t P.Value adj.P.Val B
HES6 137.79036 5.7313289 21.572562 6.400428e-41 8.372400e-37 36.66520153
DLL3 106.14625 4.3088770 19.907780 6.595993e-38 4.314110e-34 19.06034200
RGS16 118.68794 4.9943264 17.838842 5.909533e-34 2.576753e-30 24.65266014
SRRM4 133.30135 1.8829324 17.682755 1.200199e-33 3.924950e-30 10.31711616
INSM1 124.37961 1.9175950 16.849282 5.559195e-32 1.454396e-28 7.80697749
PSTPIP1 120.58200 1.2632228 16.777183 7.778568e-32 1.695857e-28 5.80279816
CBFA2T2 78.54404 4.3140507 16.615207 1.658373e-31 3.099025e-28 10.47993920
NHLH1 144.63398 2.4191008 16.173774 1.327261e-30 2.170238e-27 14.33899449
NEUROG1 182.46252 1.7460017 15.783740 8.507417e-30 1.199075e-26 17.45748702
FAM110A 103.63326 4.1012229 15.768156 9.166537e-30 1.199075e-26 13.35966764
NEUROD4 153.79389 1.0191708 14.875452 6.914986e-28 8.223176e-25 8.88826045
SSTR2 105.53379 3.4229741 14.787729 1.062928e-27 1.158680e-24 10.84231901
ELAVL3 120.97024 2.1343713 14.558902 3.276134e-27 3.296546e-24 7.36663914
GADD45G 89.87789 5.2897434 14.531386 3.752484e-27 3.506160e-24 16.77335790
NEUROD1 159.67596 1.6536858 14.442950 5.808491e-27 5.065391e-24 12.00067309
SEPTIN4 114.57766 1.7687265 14.155182 2.421594e-26 1.979805e-23 4.63078097
KIF19 136.37895 1.3771939 13.758754 1.756514e-25 1.351586e-22 5.57631803
ELAVL2 110.44799 2.7520644 13.676437 2.656080e-25 1.930233e-22 7.91156179
TFDP2 48.43140 6.4077907 13.166614 3.492107e-24 2.404224e-21 11.89863059
CHRNA3 117.42927 1.3963072 13.074253 5.584008e-24 3.652221e-21 3.44845740
EYA2 135.97013 1.6854637 12.944092 1.083472e-23 6.749000e-21 6.30965066
CLDN5 134.00845 1.3795725 12.527914 9.111648e-23 5.417703e-20 3.61751478
CACNA1A 91.11103 3.3103615 12.449231 1.365030e-22 7.763463e-20 4.96120745
SUSD2 131.07225 1.2870609 12.344459 2.340040e-22 1.275419e-19 3.36822646
CHRNA1 134.59861 0.2572580 11.971511 1.604481e-21 8.311963e-19 1.14723887
ZBTB18 87.87361 3.1687646 11.965860 1.652099e-21 8.311963e-19 4.60210476
DCX 107.52978 2.6276693 11.698102 6.620392e-21 3.207457e-18 5.16325889
EBF2 115.03242 1.5961777 11.604852 1.074673e-20 5.020642e-18 2.61195517
PLXNA1 60.06854 4.2379731 11.468423 2.185062e-20 9.856137e-18 3.35695266
CBFA2T3 92.16034 1.8429963 11.366912 3.707202e-20 1.616464e-17 0.50323017
ADGRA1 105.40870 0.8505363 10.841656 5.755227e-19 2.428520e-16 -0.32796806
IGFBPL1 92.38602 3.9587682 10.785613 7.715860e-19 3.154099e-16 6.76831057
FAM189A1 103.61750 0.4404570 10.732108 1.020877e-18 4.046695e-16 -0.88522790
MYCL 90.78804 2.8830861 10.688653 1.281581e-18 4.930694e-16 2.37783187
KIF1A 80.35540 3.9084460 9.818616 1.221861e-16 4.566617e-14 3.30445636
SH2D5 95.65907 0.7321807 9.242955 2.476931e-15 9.000206e-13 -1.60347905
RASGEF1B 73.42071 2.9083221 9.174861 3.532610e-15 1.248921e-12 0.24750260
TAGLN3 101.27309 3.8492674 9.144134 4.146037e-15 1.427219e-12 5.38671194
SYP 96.44702 1.8687858 9.025861 7.674561e-15 2.574127e-12 -0.32444743
UBE2L6 68.91058 4.9988187 9.005012 8.553695e-15 2.797272e-12 4.94041924
TFAP2C 97.94158 2.4365566 8.835238 2.066244e-14 6.592326e-12 1.13693308
CHRNB4 93.49576 0.2301937 8.760924 3.037632e-14 9.460778e-12 -1.90929720
UNCX 102.25161 0.8871815 8.676436 4.704896e-14 1.431273e-11 -1.22774447
PCBP4 79.97810 5.4426251 8.526336 1.021922e-13 3.022877e-11 6.32789589
DCC 105.98570 2.0332127 8.522956 1.039901e-13 3.022877e-11 0.59071391
TUBB2B 39.21269 8.1127821 8.483002 1.277876e-13 3.614259e-11 8.88169868
SPSB4 76.83020 3.1235051 8.479881 1.298602e-13 3.614259e-11 -0.01000446
PHF21B 73.47780 2.8678129 8.438064 1.610875e-13 4.389970e-11 -0.57735478
RAPGEF5 75.11014 2.4265624 8.433094 1.652649e-13 4.411899e-11 -0.78903859
CDH15 106.92305 1.0108015 8.403748 1.922201e-13 5.028863e-11 -1.34821426
$Cardiomyocytes
logFC AveExpr t P.Value adj.P.Val B
MYL7 130.66430 3.0976950 30.21845 1.847703e-54 2.416980e-50 46.30499
BMP4 100.17794 4.6916506 26.04480 2.482904e-48 1.244905e-44 47.33820
MYL4 128.60340 3.2151048 26.00579 2.855068e-48 1.244905e-44 43.38726
CYB5D1 83.34643 3.8300429 25.22105 4.896782e-47 1.601370e-43 38.70436
APOBEC3G 90.85741 2.0962458 23.72382 1.325661e-44 3.240680e-41 27.97143
DOK4 71.25664 4.8213956 23.69388 1.486437e-44 3.240680e-41 41.78124
CFC1 100.08962 1.1041893 22.03796 9.765033e-42 1.824806e-38 23.39502
HS3ST3A1 86.60612 2.1116755 21.92962 1.509262e-41 2.467832e-38 26.51592
GLIPR2 73.08017 3.9350171 21.68732 4.016454e-41 5.837692e-38 35.41663
BAG3 58.11844 4.2447476 21.03112 5.891112e-40 7.706163e-37 33.46104
FGF19 107.72012 2.2553657 20.82809 1.366447e-39 1.624954e-36 29.36165
BMPER 96.84503 2.3372969 20.16516 2.207314e-38 2.406156e-35 28.62389
TNNT2 92.88780 2.9350416 20.11749 2.701837e-38 2.718672e-35 30.65986
COBLL1 66.26504 3.8078148 18.55705 2.360551e-35 2.205597e-32 28.89878
DNAH2 87.33649 1.5861408 18.34031 6.195401e-35 5.402803e-32 20.95438
KRT19 65.97059 8.6797897 18.26668 8.610261e-35 7.039426e-32 49.52404
PLPPR5 84.77348 1.3670778 18.18679 1.231576e-34 9.476617e-32 18.83396
TMOD1 83.64226 1.9293714 18.15105 1.445832e-34 1.050718e-31 21.66704
PSKH2 102.71186 0.5888992 18.12316 1.638795e-34 1.078437e-31 18.74104
APLNR 121.89089 2.3444355 18.12179 1.648860e-34 1.078437e-31 29.79227
TMEM185A 53.09470 3.9425363 18.02878 2.505759e-34 1.506125e-31 27.13528
TNNI1 121.51287 3.9575031 18.02637 2.533044e-34 1.506125e-31 35.69392
RGS13 144.64986 2.6808708 17.99972 2.856335e-34 1.568971e-31 32.12976
HTRA1 80.66431 3.5987240 17.99800 2.878626e-34 1.568971e-31 29.75697
PRDM6 81.90209 1.8387960 17.78189 7.650104e-34 4.002840e-31 20.50841
LHFPL2 51.42098 3.8578678 17.43567 3.707501e-33 1.865301e-30 25.51257
ALPK2 86.60294 2.0494678 17.39412 4.485349e-33 2.173068e-30 22.24025
HAND1 134.92284 4.6368572 17.30063 6.890111e-33 3.218912e-30 37.49295
LYN 58.36667 3.5424557 17.05154 2.174038e-32 9.806410e-30 24.28785
SVEP1 71.29496 2.2052858 16.82727 6.159146e-32 2.685593e-29 19.22263
BAMBI 74.02825 6.5277939 16.79757 7.073195e-32 2.984660e-29 39.16688
SLC9A3R1 45.81782 6.6814823 16.76133 8.375453e-32 3.423728e-29 36.82614
HAS2 87.30497 5.5998335 16.66846 1.292503e-31 5.102493e-29 36.31877
ACTC1 112.70701 5.6095105 16.66295 1.326235e-31 5.102493e-29 40.55194
FABP5 58.77926 7.8679014 16.54164 2.341450e-31 8.751003e-29 44.09118
S100A11 45.05607 10.0171268 16.52148 2.573913e-31 9.352597e-29 48.96027
SHISAL2B 73.73926 2.6002339 16.10512 1.838141e-30 6.498575e-28 20.38247
TCF21 85.51477 1.8131395 15.90283 4.814762e-30 1.657419e-27 18.99167
FAM89A 76.32724 5.8430913 15.81296 7.397133e-30 2.481074e-27 36.12206
GYPB 109.95160 0.4211041 15.66734 1.486511e-29 4.861262e-27 16.12666
PPFIBP1 40.37845 5.1133169 15.50954 3.175936e-29 1.013279e-26 27.98998
TUBB6 57.58937 5.2111883 15.47312 3.785710e-29 1.179069e-26 29.67380
ADAM19 68.91500 3.7111987 15.43315 4.591396e-29 1.396745e-26 24.78315
ADGRA2 71.74334 2.0801881 15.35020 6.856677e-29 2.036366e-26 17.12242
HAPLN1 135.31468 5.1626583 15.34577 7.005309e-29 2.036366e-26 35.92405
MIXL1 109.03120 2.2806485 15.11656 2.131129e-28 6.060284e-26 22.94231
P3H2 58.24763 3.7355420 14.89919 6.156338e-28 1.713427e-25 22.47444
RGS5 89.58711 5.7892420 14.89128 6.399314e-28 1.743946e-25 33.65425
KDR 86.16713 2.9812211 14.69530 1.673609e-27 4.467853e-25 22.30438
HOXB6 110.90093 2.6245312 14.68790 1.735630e-27 4.540755e-25 24.16276
$`Ciliated epithelial cells`
logFC AveExpr t P.Value adj.P.Val B
CCNO 163.23030 1.34197803 28.29687 1.005253e-51 1.314971e-47 31.826614
CFAP126 157.20135 1.28110739 22.91650 3.006389e-43 1.966329e-39 26.502261
C11orf88 123.24933 0.46093334 19.25926 1.078980e-36 4.704714e-33 12.773345
CDC20B 123.27765 0.91970265 18.98852 3.519398e-36 1.150931e-32 17.650908
MLF1 55.05400 5.48825432 17.85601 5.467436e-34 1.430391e-30 26.357689
TEKT1 110.48383 0.16108534 17.20423 1.073871e-32 2.341217e-29 7.676046
RSPH1 107.49748 1.15679651 16.86721 5.114186e-32 9.556952e-29 11.343925
KIF9 65.33544 4.06698819 16.56006 2.147585e-31 3.511570e-28 18.282823
ROPN1L 94.85894 0.52961491 16.34023 6.041077e-31 8.780369e-28 6.491970
C1orf189 97.43277 0.31886971 16.11162 1.782348e-30 2.170743e-27 6.356314
SOX2 87.04185 4.91896284 16.10659 1.825409e-30 2.170743e-27 26.622024
FAM183A 99.33717 1.54378909 15.95431 3.766610e-30 4.105919e-27 11.991038
FOXJ1 104.97174 2.23512164 15.89446 5.011113e-30 5.042336e-27 15.726576
MUC12 80.37503 3.00255390 15.80142 7.817328e-30 7.304176e-27 15.007015
SPEF1 95.35842 0.75877560 15.66342 1.514734e-29 1.320949e-26 7.427261
C1orf194 120.85931 0.98385779 15.46962 3.850163e-29 3.147749e-26 12.530443
CCDC39 95.41475 0.66244041 14.99746 3.808497e-28 2.930526e-25 6.777590
MAP3K19 94.07040 -0.01846654 14.70544 1.592225e-27 1.157105e-24 4.116229
CAPSL 101.38635 0.29807548 14.60015 2.673245e-27 1.820235e-24 5.314272
PIFO 87.40241 3.52882807 14.59199 2.783022e-27 1.820235e-24 16.545988
CFAP52 98.63289 0.23664402 14.49739 4.438290e-27 2.764632e-24 5.085500
C9orf24 97.31268 1.05337208 13.97438 5.965985e-26 3.547320e-23 6.942407
SFRP2 74.81157 4.98786079 13.57246 4.482570e-25 2.549413e-22 20.428848
TPPP3 109.06701 2.54630137 13.46312 7.780949e-25 4.240941e-22 15.177708
FAM81B 107.60643 0.48102623 13.35301 1.357530e-24 7.103139e-22 6.193938
SPAG1 77.97744 2.54006124 12.94140 1.098461e-23 5.526528e-21 8.760483
CFAP45 106.88074 0.76783517 12.89760 1.373436e-23 6.654044e-21 6.654259
SPAG6 106.34559 0.76710346 12.45053 1.355917e-22 6.334553e-20 5.861414
C20orf85 96.91822 -0.03038045 12.35423 2.225274e-22 1.003752e-19 3.419779
ZMYND10 104.00875 0.99718113 12.29681 2.990935e-22 1.304147e-19 6.610048
SPAG8 87.20679 0.73978223 12.20234 4.867666e-22 2.053998e-19 3.698165
DAW1 84.03744 0.39915960 11.75468 4.935570e-21 2.017569e-18 2.294866
PLEKHA5 35.56665 6.47976484 11.72958 5.622365e-21 2.228671e-18 17.407452
SPAG17 82.38803 1.51749551 11.71134 6.180585e-21 2.377889e-18 5.087372
CCDC170 76.61496 0.62621966 11.56523 1.320524e-20 4.935365e-18 2.156678
RFX3 50.81374 4.23170857 11.55051 1.425561e-20 5.179934e-18 10.015639
C5orf49 101.88563 1.94581738 11.52209 1.652612e-20 5.842655e-18 9.088395
TPBG 91.29992 6.17059639 11.27743 5.910331e-20 2.034554e-17 21.753149
RSPH4A 99.14527 1.04673557 11.19386 9.139565e-20 3.065504e-17 5.628114
MCUB 65.42028 3.61777699 11.15422 1.123974e-19 3.675677e-17 8.240501
CCDC114 82.49538 0.38231633 11.04939 1.943020e-19 6.199181e-17 1.662925
DRC1 107.58455 0.80926145 10.83577 5.935310e-19 1.848567e-16 4.826680
IGFBPL1 69.22922 3.95876817 10.74157 9.715524e-19 2.955553e-16 11.661006
WDR49 91.96656 0.44017013 10.66534 1.447915e-18 4.304584e-16 2.605762
AKAP14 80.99571 0.27168377 10.56856 2.403258e-18 6.986003e-16 1.365864
LRRC73 72.98617 1.68691586 10.42588 5.074221e-18 1.442954e-15 2.994006
PLXDC2 53.37388 3.92130700 10.41727 5.308163e-18 1.477363e-15 7.161856
DNAH7 85.80930 0.95375866 10.36158 7.106661e-18 1.936713e-15 3.075866
EFHC1 52.83757 4.46622294 10.31434 9.102208e-18 2.429918e-15 9.754151
BMP7 47.07608 4.37485330 10.18567 1.786249e-17 4.673184e-15 7.989635
$`Ductal cells`
logFC AveExpr t P.Value adj.P.Val B
SHH 143.68142 0.7005265 24.00904 4.476841e-45 5.856156e-41 25.484185
GASK1B 111.42556 1.9409450 21.82597 2.292073e-41 1.499130e-37 26.162351
IHH 110.65252 1.5264822 20.63833 3.013624e-39 1.314041e-35 21.719632
TMEM229A 104.30223 1.2756792 19.34869 7.316162e-37 2.392568e-33 18.251292
FOXA2 114.43579 2.3626344 19.23913 1.177750e-36 3.081229e-33 26.428385
MNX1 105.60992 1.5673352 18.50659 2.953525e-35 6.439177e-32 19.380783
VIL1 87.36363 2.0194712 18.42958 4.160683e-35 7.775128e-32 18.740219
CST1 167.14941 3.0632741 18.14218 1.504589e-34 2.460192e-31 33.779978
CST4 129.03536 1.5048488 17.79236 7.295442e-34 1.060352e-30 23.122807
HPGD 92.90406 2.5690598 17.02789 2.425680e-32 3.173032e-29 22.107840
RAB25 86.89581 3.8125574 16.98471 2.963137e-32 3.523708e-29 27.763366
FOXA1 96.01516 1.2318728 16.80737 6.757564e-32 7.366308e-29 15.085899
SCGB1A1 102.34758 0.1694337 16.59235 1.845871e-31 1.857372e-28 13.048433
FOXQ1 106.89928 2.0403927 16.43284 3.904432e-31 3.648134e-28 19.755407
WFDC2 72.85817 5.5491577 16.02459 2.695232e-30 2.350422e-27 32.892269
ALPL 66.50418 3.8378051 15.72366 1.134488e-29 9.275149e-27 22.147217
KLF5 73.47058 2.4044719 15.53447 2.816366e-29 2.167111e-26 16.140863
CLDN3 70.70027 3.3835805 15.17622 1.594377e-28 1.158669e-25 20.649345
NENF 80.20328 6.4620381 14.69911 1.642583e-27 1.130875e-24 34.223838
SMAGP 57.94867 3.4949297 14.59426 2.752014e-27 1.799955e-24 17.329190
CAPN9 95.35551 1.0291877 14.36429 8.573447e-27 5.340441e-24 11.712069
CDH17 89.86364 0.1431509 14.27014 1.367541e-26 8.131274e-24 7.208636
IGFBP6 89.25511 2.7075884 14.21563 1.792784e-26 1.019626e-23 18.687988
ZFP36L2 56.75904 6.5312268 14.19452 1.991200e-26 1.085287e-23 30.061186
ARHGEF19 55.80740 2.8774848 14.09315 3.298280e-26 1.725491e-23 14.028964
SLC12A2 50.63194 4.6590684 14.08531 3.429614e-26 1.725491e-23 21.783638
EPCAM 96.33657 5.6005879 14.01170 4.951503e-26 2.398912e-23 29.962477
ANXA3 76.35277 4.3416845 13.97558 5.930321e-26 2.770519e-23 23.446413
CLDN4 92.06373 2.9275341 13.96752 6.173848e-26 2.784831e-23 20.501571
SOX17 132.40726 2.5971138 13.95660 6.520269e-26 2.843055e-23 21.945171
KRT18 59.37421 8.5293745 13.92361 7.689691e-26 3.243472e-23 35.272731
DSP 56.27506 7.0748749 13.91734 7.934493e-26 3.243472e-23 31.084035
ECHDC1 32.50908 6.5067194 13.82183 1.280131e-25 5.074363e-23 26.964859
TNNC1 78.28109 4.0471838 13.77244 1.639945e-25 6.309447e-23 22.462253
TRIM17 78.80793 2.2665971 13.69481 2.421820e-25 9.051379e-23 13.310627
KITLG 77.52114 2.8648700 13.58284 4.254059e-25 1.545760e-22 14.802235
C10orf82 60.12093 2.9468109 13.57226 4.486955e-25 1.586320e-22 14.369047
SERPINI1 60.40501 3.1162122 13.48031 7.134108e-25 2.455823e-22 14.076346
FREM2 61.54915 3.9451632 13.46815 7.585900e-25 2.544389e-22 19.039074
GJA4 86.58513 1.4068779 13.36670 1.266669e-24 4.142326e-22 10.562944
F2 75.96756 1.0658412 13.19079 3.088712e-24 9.854499e-22 7.941239
SAT1 62.05157 6.3992827 13.18354 3.204519e-24 9.980550e-22 28.732950
SGK1 78.82008 2.0537412 13.04839 6.369178e-24 1.937563e-21 12.001188
SH3YL1 41.29040 4.4045214 12.92805 1.175834e-23 3.495702e-21 17.556978
APOA2 87.99515 4.5700960 12.82426 1.997231e-23 5.805727e-21 21.578721
TMEM54 54.18468 3.5830793 12.80886 2.160715e-23 6.144417e-21 15.814817
DEFA5 95.13129 0.4415613 12.77085 2.624180e-23 7.303596e-21 7.902071
FBLN2 61.42286 4.0725440 12.71361 3.516942e-23 9.584400e-21 17.632106
PCBD1 33.40965 6.9252945 12.64991 4.873590e-23 1.301050e-20 25.673380
SHISAL2B 66.63326 2.6002339 12.57839 7.032133e-23 1.839747e-20 12.248142
$`Epicardial fat cells`
logFC AveExpr t P.Value adj.P.Val B
SLC9A3R1 94.32841 6.6814823 31.17107 9.138907e-56 1.195460e-51 49.580065
AQP1 164.03920 1.8756794 29.33866 3.175444e-53 2.076899e-49 31.780688
COBLL1 110.26270 3.8078148 28.06091 2.228486e-51 9.716943e-48 32.216841
ANXA8 139.36332 0.8855355 23.88767 7.097668e-45 2.321115e-41 18.947166
TNNT2 121.16601 2.9350416 22.04344 9.552540e-42 2.499136e-38 25.527118
SERPINE3 157.28547 0.3786379 21.35077 1.582165e-40 3.449383e-37 18.580813
CARD16 130.10506 0.4869025 20.78315 1.647295e-39 3.078323e-36 13.798601
SLC40A1 120.91422 3.3828801 20.33706 1.067363e-38 1.745271e-35 26.954025
FIBIN 120.34115 1.8971410 19.80080 1.042173e-37 1.400640e-34 18.273241
SLC5A12 122.43393 2.7850221 19.79448 1.070744e-37 1.400640e-34 23.247790
SLC34A2 134.51562 0.5276665 19.63829 2.093532e-37 2.489590e-34 13.149710
FRZB 135.29174 5.4381793 19.52031 3.480986e-37 3.794565e-34 37.826990
SLN 172.16274 2.2626310 19.17227 1.575945e-36 1.585764e-33 28.166227
SARAF 47.45876 7.8538281 19.12684 1.921526e-36 1.795391e-33 36.328088
CD44 105.09442 2.5102088 18.87231 5.862353e-36 5.112363e-33 17.213029
KRT24 125.83223 0.6689462 18.37455 5.317432e-35 4.347333e-32 12.817443
SLC7A7 113.14202 2.5859236 18.28548 7.915664e-35 5.756301e-32 19.029768
LIMS1 50.07334 6.2651233 18.28534 7.920909e-35 5.756301e-32 27.400972
MYRF 91.47066 3.1829695 17.66979 1.273151e-33 8.765307e-31 17.221080
NPNT 112.80533 2.4641667 17.50080 2.751941e-33 1.799907e-30 17.464960
ATP7B 89.64066 3.0250264 17.47059 3.159827e-33 1.968271e-30 15.340508
COL1A1 115.44418 6.2764999 17.25933 8.331386e-33 4.953767e-30 34.605028
MYL4 115.74911 3.2151048 17.22731 9.655107e-33 5.491237e-30 22.472821
AKR1B15 112.24352 1.5067348 17.00306 2.721502e-32 1.483332e-29 12.536939
RAMP1 104.13799 3.0053905 16.95251 3.440539e-32 1.767709e-29 18.412173
LUM 152.36937 4.7155844 16.94799 3.513525e-32 1.767709e-29 31.194031
AEBP1 66.19705 4.4020690 16.70945 1.067099e-31 5.169897e-29 17.839711
DSC3 96.44074 1.9939554 16.56494 2.098997e-31 9.806063e-29 10.997464
BNC1 109.40344 1.2420685 16.47173 3.251676e-31 1.466730e-28 10.605918
IFIT1 97.15666 3.0191987 16.33195 6.281854e-31 2.739098e-28 16.400664
STX18 53.61765 5.5193489 16.31759 6.722296e-31 2.836592e-28 21.996926
DCN 170.29764 3.1437291 16.24605 9.426333e-31 3.853308e-28 26.585083
PRCD 101.98930 1.8232319 16.03631 2.549072e-30 1.010437e-27 10.994048
PRSS35 104.46662 3.2397555 15.76657 9.236339e-30 3.553546e-27 17.902110
TGFBR3 83.13219 3.1869881 15.66822 1.480237e-29 5.401942e-27 14.315488
THBD 109.86076 1.2038102 15.66732 1.486659e-29 5.401942e-27 9.497693
CYB5R2 71.65741 3.6845580 15.44968 4.239161e-29 1.498715e-26 14.138403
FMO1 121.00192 1.8281981 15.43197 4.617548e-29 1.589530e-26 13.891087
TNFAIP3 83.59592 2.5601386 15.42171 4.852378e-29 1.627537e-26 11.006959
LEPROTL1 46.89480 6.6570553 15.35822 6.595722e-29 2.156966e-26 24.997326
COL8A2 113.61595 0.8062160 15.20363 1.395583e-28 4.452591e-26 8.202056
CREB3L1 91.05316 3.4812380 15.17808 1.580030e-28 4.921040e-26 15.947947
COL21A1 106.65926 2.4613167 15.11512 2.146139e-28 6.528756e-26 13.909096
DDR2 99.44669 2.8971187 14.95038 4.793167e-28 1.424987e-25 14.809412
BNC2 90.04942 3.2479340 14.77729 1.118775e-27 3.252155e-25 14.694171
COL5A1 89.20959 4.4965267 14.68603 1.751697e-27 4.981292e-25 20.371664
COL3A1 145.76480 5.3883411 14.50923 4.186286e-27 1.165124e-24 28.085792
POSTN 144.74872 3.8513366 14.46962 5.091088e-27 1.387428e-24 23.648476
GALNT10 71.67659 3.8606737 14.29188 1.227626e-26 3.277261e-24 14.009655
MORC4 61.06956 3.6446214 14.19274 2.008873e-26 5.255613e-24 10.606694
$`Goblet cells`
logFC AveExpr t P.Value adj.P.Val B
CCKBR 110.06733 2.8864382 27.13638 5.299746e-50 6.932597e-46 34.45488
FAM184A 111.18722 3.7029690 26.59280 3.547390e-49 2.320171e-45 39.16682
CCDC81 112.89029 2.0075884 24.57342 5.362603e-46 2.338274e-42 28.75924
FOXA2 135.58586 2.3626344 24.36400 1.173844e-45 3.838765e-42 33.43206
PLSCR2 156.23423 1.2118813 22.46276 1.794525e-42 4.694836e-39 30.53716
FOXQ1 127.32312 2.0403927 22.17410 5.660983e-42 1.234189e-38 29.48584
EPSTI1 90.64317 3.4167152 21.75971 2.995958e-41 5.598589e-38 30.13592
PRSS1 131.85935 0.8334319 21.55212 6.955368e-41 1.137290e-37 23.75502
WFDC2 94.70063 5.5491577 21.13068 3.906486e-40 5.677861e-37 42.06517
PDZK1 103.45192 1.2293187 20.59803 3.566782e-39 4.665707e-36 19.35213
HHEX 122.18081 1.3410679 19.98705 4.704070e-38 5.593995e-35 23.16529
MNX1 107.41996 1.5673352 19.74719 1.311325e-37 1.429454e-34 21.93046
CST1 179.17866 3.0632741 19.60511 2.414928e-37 2.429975e-34 36.22476
PCBD1 50.64427 6.9252945 19.25038 1.121471e-36 1.047855e-33 39.82232
CER1 127.15289 2.0397658 19.22920 1.229750e-36 1.072424e-33 26.75497
APOA2 119.66504 4.5700960 19.20541 1.363996e-36 1.115152e-33 35.90340
ADM 92.94778 2.6525145 18.53301 2.626421e-35 2.020954e-32 23.25370
COCH 78.55069 2.9374724 18.51580 2.835125e-35 2.060348e-32 21.82873
VIL1 87.76423 2.0194712 18.00399 2.801959e-34 1.916170e-31 17.55428
PRSS2 147.93660 1.1202828 17.99410 2.929700e-34 1.916170e-31 24.34888
LGR5 100.62783 2.0058344 17.85650 5.455356e-34 3.398167e-31 20.82498
HP 91.12315 0.7621428 17.23865 9.163674e-33 5.448637e-30 12.36840
PRDM1 86.35157 2.2010891 16.94614 3.543894e-32 2.015551e-29 17.77801
LAPTM4B 41.72772 8.8668685 16.83053 6.066454e-32 3.306470e-29 42.72303
SEMA3E 99.20815 2.3329429 16.72611 9.871974e-32 5.165412e-29 21.32080
GSC 114.41262 1.2115720 16.69491 1.142154e-31 5.746352e-29 18.44214
TRDN 116.24882 2.4302318 16.62183 1.607734e-31 7.789176e-29 23.68620
GATA4 89.42945 2.7684350 16.61108 1.690749e-31 7.898819e-29 21.20670
APOC1 67.04166 6.5875047 16.41397 4.267182e-31 1.924794e-28 34.17720
P3H2 68.58017 3.7355420 16.35614 5.604186e-31 2.443612e-28 22.42240
PIK3R5 98.47909 1.0060652 16.20222 1.159931e-30 4.894535e-28 13.88334
SLC2A3 78.14671 6.2843433 15.97168 3.467387e-30 1.417403e-27 33.92845
CARHSP1 30.78005 7.6020921 15.92244 4.384780e-30 1.738100e-27 33.87210
SOX17 144.19265 2.5971138 15.72429 1.131055e-29 4.351569e-27 25.92978
FZD5 124.92947 2.5612493 15.60152 2.039471e-29 7.622378e-27 23.92463
PTGR1 63.85621 7.4625797 15.35187 6.801564e-29 2.471424e-26 36.45198
IHH 92.79370 1.5264822 15.29866 8.800670e-29 3.111393e-26 13.79140
RAB25 84.74324 3.8125574 15.28362 9.466381e-29 3.258677e-26 22.72676
SHISAL2B 77.09117 2.6002339 15.21107 1.346002e-28 4.514628e-26 16.47193
KCNK12 92.47674 2.3998878 15.11194 2.179672e-28 7.128072e-26 18.01654
PLCXD3 78.63658 1.3469952 15.06235 2.775100e-28 8.853922e-26 10.88065
S100A16 72.61461 4.6850373 14.47149 5.044063e-27 1.544670e-24 23.35201
VAMP8 63.76696 5.2537599 14.47015 5.077655e-27 1.544670e-24 24.95638
SLCO2A1 78.39423 1.7276243 14.10620 3.090593e-26 9.188192e-24 11.76974
DMKN 72.43869 6.0650201 13.99308 5.433838e-26 1.579556e-23 27.86286
TFF3 106.42073 1.2060964 13.88830 9.175538e-26 2.609244e-23 13.06430
NRG1 67.79722 3.1473479 13.83526 1.196784e-25 3.330879e-23 16.57545
SERPINE2 76.13986 6.3636276 13.77987 1.579945e-25 4.305679e-23 28.51404
DENND2C 81.87393 2.2918567 13.71887 2.145970e-25 5.728865e-23 13.85858
FOXA1 86.35555 1.2318728 13.69756 2.388560e-25 6.220098e-23 10.05577
$`Granule neurons`
logFC AveExpr t P.Value adj.P.Val B
ELAVL3 156.33323 2.1343713 32.36363 2.347957e-57 2.446583e-53 33.67212
LHX9 167.85888 2.0780892 32.20993 3.740666e-57 2.446583e-53 35.76887
UNCX 162.10375 0.8871815 31.87166 1.049182e-56 4.574782e-53 26.49825
INSM1 140.90935 1.9175950 30.55843 6.265015e-55 2.048816e-51 27.84146
NHLH1 177.85335 2.4191008 30.28651 1.486868e-54 3.889944e-51 39.68953
IGFBPL1 136.12734 3.9587682 27.91827 3.614750e-51 7.880757e-48 40.30392
ST18 155.66791 0.5091356 27.64646 9.133926e-51 1.664765e-47 20.29504
KLHL35 141.33031 2.1556421 27.61476 1.018127e-50 1.664765e-47 28.66000
TLCD3B 139.20300 1.7731339 27.53008 1.361277e-50 1.978541e-47 24.09398
KIF5C 112.28194 4.4306399 25.35045 3.051426e-47 3.991570e-44 36.74672
EBF3 135.22512 1.2507997 24.84783 1.934684e-46 2.300691e-43 19.04715
DLL3 98.38386 4.3088770 24.71690 3.143596e-46 3.426781e-43 33.55651
DCX 134.38971 2.6276693 24.42075 9.488791e-46 9.547914e-43 28.50277
CRMP1 112.53436 3.7555245 24.33749 1.296674e-45 1.211556e-42 30.41818
INA 114.57696 2.6686304 24.12829 2.851098e-45 2.486348e-42 23.18005
SYP 130.93550 1.8687858 24.05421 3.772801e-45 3.084500e-42 21.57818
CELF3 133.83305 1.2546600 23.83143 8.792364e-45 6.765465e-42 17.73571
TAGLN3 148.73051 3.8492674 23.60610 2.080385e-44 1.511862e-41 39.31041
DRAXIN 102.70779 3.3897111 23.27939 7.324726e-44 5.042881e-41 24.57928
OLFM1 117.16334 2.7883832 23.11455 1.388522e-43 9.081628e-41 24.84213
MAP1B 79.33089 8.7464887 22.93908 2.752263e-43 1.714398e-40 55.77921
ELAVL2 122.53480 2.7520644 22.61322 9.897444e-43 5.884930e-40 25.61333
SSTR2 109.70176 3.4229741 21.64626 4.744313e-41 2.698277e-38 26.61585
STMN2 146.68650 4.7054494 21.55954 6.748672e-41 3.678307e-38 41.85128
CNTN2 143.86429 1.5307494 21.47216 9.634296e-41 5.041049e-38 21.80347
EBF2 130.72138 1.5961777 21.44444 1.078814e-40 5.427678e-38 18.91179
INSM2 139.33855 0.3073516 21.43179 1.136007e-40 5.503743e-38 11.97642
GNG3 101.76171 2.6673015 21.14580 3.670578e-40 1.714815e-37 17.63171
DCC 141.76902 2.0332127 21.05629 5.309289e-40 2.394855e-37 23.52783
TCP10L 108.81063 1.7550309 20.97527 7.421458e-40 3.236003e-37 13.49612
CFAP298 70.70351 6.2406198 20.63205 3.093708e-39 1.305445e-36 36.00796
HES6 113.71186 5.7313289 20.46766 6.160804e-39 2.518421e-36 41.01402
RTN1 124.62127 3.1365014 20.36582 9.455307e-39 3.748027e-36 26.37755
ADCYAP1 126.32955 0.8113424 20.25883 1.485007e-38 5.713345e-36 12.09939
TUBB2B 60.57004 8.1127821 19.98654 4.714264e-38 1.761922e-35 43.28248
MLLT11 71.95415 7.5497265 18.94765 4.210281e-36 1.529853e-33 41.32164
NEUROD1 159.60363 1.6536858 18.77878 8.850720e-36 3.129088e-33 23.65809
CCNO 142.98429 1.3419780 18.71684 1.163368e-35 4.004742e-33 16.12188
NRXN1 116.52061 2.7210766 18.34759 5.997303e-35 2.011557e-32 19.62318
SCG3 125.34658 2.9950166 18.16603 1.351788e-34 4.420684e-32 23.41934
RGS16 100.52619 4.9943264 17.95143 3.551671e-34 1.133156e-31 30.80409
TBR1 150.83697 0.6804992 17.81831 6.485756e-34 2.020004e-31 14.94161
NHLH2 104.28010 1.9647414 17.25003 8.696093e-33 2.645432e-30 11.73556
CALY 104.26306 2.8062889 17.19998 1.095136e-32 3.255789e-30 16.53923
NPTX2 113.24127 1.6393260 17.15161 1.368920e-32 3.979299e-30 10.93535
SRRM4 110.62813 1.8829324 16.95499 3.401226e-32 9.672051e-30 13.43849
RORB 127.85261 1.2607454 16.78292 7.573349e-32 2.107808e-29 11.94338
FNDC5 100.77977 2.9112692 16.76738 8.142510e-32 2.219004e-29 16.37960
RAB26 104.73355 1.9711739 16.69505 1.141405e-31 3.047086e-29 11.38274
CACNA1A 87.09858 3.3103615 16.66791 1.295838e-31 3.390171e-29 15.27027
$Hepatoblasts
logFC AveExpr t P.Value adj.P.Val B
APOB 212.86304 2.418662449 68.74141 6.875979e-91 8.994469e-87 58.83317
RBP4 240.42874 2.706134932 65.40843 1.291711e-88 6.583320e-85 63.74390
AHSG 236.55900 1.178939669 65.31151 1.509820e-88 6.583320e-85 53.12502
AFP 239.64321 2.782945774 64.82664 3.306424e-88 1.081283e-84 63.19406
FGA 220.48744 1.156850499 62.39136 1.847634e-86 4.833781e-83 51.18439
APOC3 243.09001 2.002620895 57.22719 1.568987e-82 3.420652e-79 57.89520
SMLR1 178.81594 1.189693142 57.11940 1.910453e-82 3.570091e-79 45.63108
GSTA2 193.54753 1.867321844 56.98326 2.451045e-82 4.007766e-79 51.72488
FGG 224.47363 1.260810569 55.93150 1.712504e-81 2.489030e-78 51.28503
FGB 236.11890 1.690912416 53.10761 3.766354e-79 4.926768e-76 54.68095
APOM 163.46578 3.604052826 53.04605 4.248771e-79 5.052561e-76 59.21709
G0S2 208.17758 2.071547317 51.92153 3.929938e-78 4.283960e-75 54.20816
SERPINA1 225.53980 0.926589202 51.80391 4.972186e-78 5.003166e-75 49.84857
LGALS2 168.44205 0.845959810 48.62786 3.462019e-75 3.084435e-72 40.53230
APOA2 234.91118 4.570095953 48.61777 3.536926e-75 3.084435e-72 69.87807
FABP1 226.38227 1.395422395 48.00378 1.311769e-74 1.072453e-71 50.80848
AGT 174.29327 1.272051385 46.57228 2.959215e-73 2.277029e-70 44.10895
MEP1A 153.38844 0.518379947 45.38133 4.227896e-72 3.072506e-69 34.98358
DPYS 167.01021 0.792165370 45.05930 8.772404e-72 6.039569e-69 39.27715
GLTPD2 145.26622 0.850389220 44.29380 5.068883e-71 3.315303e-68 35.69155
GLUD1 96.74332 5.297080823 44.05679 8.773384e-71 5.464982e-68 59.73280
ASGR2 148.70094 1.089839566 43.50328 3.192191e-70 1.898048e-67 37.76862
MTTP 176.22017 1.291211877 43.35006 4.576000e-70 2.602551e-67 43.06298
DGAT2 125.77295 2.373470168 41.45300 4.352831e-68 2.372474e-65 40.97247
CREB3L3 145.24734 0.408790423 40.81372 2.106213e-67 1.068196e-64 32.18688
ARG1 155.47565 0.198744047 40.81049 2.123163e-67 1.068196e-64 32.94283
APOC1 124.15618 6.587504705 39.87575 2.215231e-66 1.073238e-63 73.35468
AKR1D1 153.81489 0.861641965 39.77276 2.876707e-66 1.343936e-63 36.22255
F2 143.97979 1.065841190 39.72012 3.288511e-66 1.483345e-63 35.75914
SLC39A5 141.60591 0.875527933 39.55897 4.957895e-66 2.161807e-63 33.80627
GPD1 138.49176 0.643634062 39.17875 1.313719e-65 5.543470e-63 31.14492
PRODH2 145.96368 0.336214318 38.73797 4.107670e-65 1.679138e-62 31.79756
AMBP 152.55830 1.117420768 38.65097 5.151042e-65 2.041842e-62 36.91930
ACAA1 82.06253 5.203223501 38.22066 1.588295e-64 6.110731e-62 53.90432
DUSP9 135.92510 1.779731417 37.37224 1.510794e-63 5.646484e-61 37.35442
CEBPA 119.73610 2.361272139 36.34215 2.471104e-62 8.979032e-60 38.22406
SULT1E1 144.65131 -0.004876959 36.14546 4.245791e-62 1.501059e-59 28.71980
GOLT1A 142.67281 1.353479076 35.78202 1.161998e-61 4.000025e-59 35.65822
A1CF 137.11461 0.384271567 35.69544 1.478812e-61 4.960086e-59 29.05437
SLC13A5 137.09125 1.039275002 35.56561 2.124937e-61 6.949074e-59 32.23707
KLB 132.42762 0.706396763 35.53696 2.302242e-61 7.345276e-59 29.30103
SULT2A1 139.06381 0.034109509 35.34683 3.923963e-61 1.222128e-58 26.60808
MPC2 68.41584 7.768511280 35.23282 5.408648e-61 1.645361e-58 71.78758
HNF4A 139.29951 1.216225955 35.08876 8.123476e-61 2.415072e-58 34.85642
GSTA1 172.21517 1.615372085 35.05892 8.838985e-61 2.569395e-58 42.28580
HP 133.73989 0.762142835 34.69744 2.469843e-60 7.023482e-58 30.71709
PLA2G12B 127.67459 1.011311146 34.13228 1.253855e-59 3.489719e-57 30.42250
HPX 137.23671 1.382479304 33.89421 2.502568e-59 6.820020e-57 35.37624
HMGCS2 157.06968 0.987059704 33.82302 3.079407e-59 8.220760e-57 36.17443
DPP4 137.60063 2.059792389 33.65851 4.980269e-59 1.302938e-56 39.17974
$IG
logFC AveExpr t P.Value adj.P.Val B
PCOLCE 107.35012 4.8474419 22.99736 2.192014e-43 2.867374e-39 40.332385
COL1A1 127.72961 6.2764999 22.78231 5.086897e-43 3.327085e-39 50.418184
COL6A2 101.54411 6.2693029 21.43958 1.100416e-40 4.798182e-37 46.113122
COL6A3 131.16234 3.4290288 21.35557 1.551376e-40 5.073386e-37 33.865361
EMILIN1 106.27719 2.8739884 20.58518 3.763778e-39 9.846797e-36 25.467439
TWIST1 116.52184 3.0945849 20.17875 2.083820e-38 4.543075e-35 29.401862
KCTD12 99.51344 3.9630040 18.55580 2.373661e-35 4.435694e-32 28.913021
COL3A1 159.41304 5.3883411 18.42341 4.276534e-35 6.992668e-32 40.676606
DDR2 102.38580 2.8971187 18.39317 4.893657e-35 7.112659e-32 23.623001
COL6A1 72.85261 6.8556861 17.37838 4.820957e-33 6.306294e-30 38.431250
LUM 142.64713 4.7155844 17.13212 1.497838e-32 1.781202e-29 34.084319
COL21A1 102.18611 2.4613167 16.77456 7.874391e-32 8.583742e-29 19.636445
COL1A2 94.94535 6.9908439 16.33419 6.215577e-31 6.254305e-28 39.190941
IGFBP3 122.97875 4.8257654 16.31244 6.887631e-31 6.364335e-28 32.281036
TCF21 102.56134 1.8131395 16.29913 7.334614e-31 6.364335e-28 14.967569
DCN 157.72513 3.1437291 16.28653 7.784524e-31 6.364335e-28 29.185572
PCDH18 71.58878 3.4732442 16.15493 1.451307e-30 1.116738e-27 17.514833
MFAP4 106.23234 4.0818825 16.11096 1.787905e-30 1.299310e-27 27.192346
COL5A1 84.98473 4.4965267 15.91789 4.480961e-30 3.085023e-27 26.072110
NID2 93.61667 3.8234636 15.86680 5.718682e-30 3.740304e-27 23.644215
CD248 92.47414 2.2968251 15.67554 1.429097e-29 8.901911e-27 14.494484
FIBIN 99.02207 1.8971410 15.36795 6.292509e-29 3.741468e-26 13.300779
PRRX2 80.18279 2.7710212 15.13155 1.981196e-28 1.126784e-25 13.734089
S1PR3 71.21109 3.7477246 15.02896 3.265870e-28 1.780035e-25 17.523437
CPED1 102.85334 2.0788448 14.92893 5.322870e-28 2.785138e-25 15.659027
CTHRC1 78.80884 4.8363794 14.81482 9.306949e-28 4.682469e-25 24.389212
POSTN 133.39569 3.8513366 14.58473 2.884441e-27 1.397458e-24 26.384484
HAND2 116.48710 2.7539369 14.31021 1.120933e-26 5.236760e-24 20.112630
EMP3 63.75094 5.6615425 14.15489 2.425144e-26 1.093907e-23 25.058909
TNNT2 85.05714 2.9350416 14.05066 4.076493e-26 1.777487e-23 15.103544
IFITM2 54.40575 5.5045974 13.99848 5.289394e-26 2.231953e-23 22.043856
ADGRA2 82.60776 2.0801881 13.93611 7.223507e-26 2.952834e-23 9.951716
PPIC 66.33688 5.7541758 13.77488 1.619989e-25 6.421539e-23 24.454184
PTH1R 81.86709 3.1660453 13.75780 1.764965e-25 6.610450e-23 15.399778
COL5A2 75.14705 5.1467724 13.75737 1.768716e-25 6.610450e-23 23.823467
HOXA13 102.49839 0.6432163 13.70643 2.284354e-25 8.300454e-23 7.279515
HGF 108.53089 1.0148773 13.61502 3.617731e-25 1.279015e-22 9.882073
TNNI1 108.76895 3.9575031 13.59767 3.947843e-25 1.358993e-22 22.393362
GPX8 42.24953 6.1566286 13.54314 5.196326e-25 1.742901e-22 22.636721
LGALS1 68.69366 6.8922090 13.36873 1.253724e-24 4.099990e-22 28.444800
ADAMTS1 68.57596 4.2288417 13.33013 1.524205e-24 4.862957e-22 17.479055
IGFBP4 55.21606 4.6457332 13.26680 2.100660e-24 6.542556e-22 16.569742
DACT3 62.13326 3.1881491 13.21989 2.664765e-24 8.106463e-22 10.934974
IL11RA 77.92492 3.4666733 12.83003 1.939196e-23 5.765142e-21 14.518390
PTGFR 85.70433 0.2592682 12.55070 8.105967e-23 2.356314e-20 3.534438
CCDC178 90.53569 0.8326892 12.52316 9.336784e-23 2.653371e-20 5.620593
GLT8D2 81.33065 2.0016704 12.51910 9.533555e-23 2.653371e-20 8.154654
DSC3 73.09694 1.9939554 12.27955 3.269119e-22 8.909031e-20 7.264299
EVA1B 58.75781 5.1275367 12.17275 5.670653e-22 1.513833e-19 17.438167
RGS5 84.95501 5.7892420 12.15141 6.331176e-22 1.656362e-19 23.063368
$`Intestinal epithelial cells`
logFC AveExpr t P.Value adj.P.Val B
APOB 129.95498 2.4186624 35.11027 7.644055e-61 9.999188e-57 41.87566
APOC1 102.22306 6.5875047 31.78707 1.359773e-56 7.316419e-53 65.66334
MTTP 140.38054 1.2912119 31.71861 1.677949e-56 7.316419e-53 35.35848
HNF4A 122.75461 1.2162260 29.80563 6.960383e-54 2.276219e-50 31.38704
APOA2 150.41088 4.5700960 28.44029 6.211640e-52 1.373633e-48 53.84341
FGB 149.49898 1.6909124 28.43604 6.300589e-52 1.373633e-48 37.71880
IHH 125.90842 1.5264822 28.36886 7.892873e-52 1.474952e-48 33.85802
AKR1D1 118.24768 0.8616420 27.08390 6.359570e-50 1.039869e-46 26.82426
LGALS2 113.24136 0.8459598 26.95531 9.951871e-50 1.446449e-46 25.97447
F2 108.45780 1.0658412 26.57627 3.760203e-49 4.918721e-46 26.36877
VIL1 102.87268 2.0194712 26.06824 2.283275e-48 2.644064e-45 30.54510
APOA1 185.01295 5.3603629 26.05134 2.425561e-48 2.644064e-45 58.09812
CCKBR 100.32241 2.8864382 25.98694 3.054535e-48 3.073568e-45 36.08047
DPP4 111.16138 2.0597924 25.71711 8.063341e-48 7.534040e-45 32.79172
SLC39A5 104.45023 0.8755279 25.68532 9.044632e-48 7.887522e-45 24.12762
S100A14 151.30196 2.3619882 25.49306 1.815480e-47 1.484268e-44 39.34440
S100A16 103.08890 4.6850373 25.40173 2.531051e-47 1.947569e-44 45.87603
PDZK1 110.63115 1.2293187 25.33917 3.179687e-47 2.310749e-44 27.16589
CST4 154.22922 1.5048488 25.26060 4.237002e-47 2.917064e-44 36.47398
FGFR1 51.99247 6.6221206 25.07050 8.508463e-47 5.564960e-44 51.75694
ADD3 51.98290 6.1224418 24.07269 3.518046e-45 2.191407e-42 46.15169
PCBD1 54.35431 6.9252945 23.74113 1.240833e-44 7.377881e-42 52.33702
SERPINE2 106.01883 6.3636276 23.70413 1.429277e-44 8.128860e-42 55.22374
FOXA2 124.88964 2.3626344 23.61529 2.008358e-44 1.094639e-41 35.11882
CYP4X1 100.75570 1.8636082 23.49951 3.132723e-44 1.639166e-41 28.00295
GSTA2 103.91056 1.8673218 23.46481 3.580188e-44 1.801247e-41 28.04614
HABP2 108.37819 0.6195381 23.28953 7.042669e-44 3.412043e-41 22.57591
APOM 88.30447 3.6040528 23.21230 9.499097e-44 4.437774e-41 35.28271
SMLR1 98.68692 1.1896931 23.00741 2.107725e-43 9.507293e-41 22.14613
FGFR4 102.71134 2.9248053 22.96574 2.480028e-43 1.081375e-40 33.57896
RBP4 111.19146 2.7061349 22.91325 3.044810e-43 1.284812e-40 33.88021
CST1 191.38537 3.0632741 22.57981 1.129310e-42 4.616407e-40 43.19309
FOXQ1 119.78729 2.0403927 22.34083 2.911995e-42 1.154297e-39 32.45299
PLA2G12B 94.11714 1.0113111 21.68918 3.986194e-41 1.533629e-38 19.79505
FZD4 71.45670 2.5833133 20.45452 6.510352e-39 2.433198e-36 23.67553
FGA 103.23256 1.1568505 19.97099 5.037309e-38 1.830362e-35 20.80888
CHDH 70.69916 2.5362295 19.87511 7.583689e-38 2.681142e-35 22.46718
GAMT 43.56159 6.5280758 19.82255 9.494932e-38 3.268506e-35 41.92461
SLCO2A1 88.65477 1.7276243 19.63253 2.146046e-37 7.198059e-35 22.61199
DENND2C 95.43203 2.2918567 19.31508 8.465243e-37 2.768346e-34 25.48421
MEP1A 89.49524 0.5183799 19.25852 1.082424e-36 3.453459e-34 14.75312
PPFIBP2 64.77025 3.1999294 19.06787 2.486330e-36 7.743734e-34 24.82958
MGST2 65.56294 5.7152198 18.77223 9.110286e-36 2.771434e-33 38.87630
ASGR2 84.90049 1.0898396 18.70249 1.239542e-35 3.685102e-33 17.09172
RAB25 90.46080 3.8125574 18.57204 2.208640e-35 6.420272e-33 31.70362
CTSE 110.34638 0.6346451 18.46498 3.553953e-35 1.010636e-32 18.24168
MARC1 54.65659 4.2794344 18.43570 4.048757e-35 1.126847e-32 28.41843
APOC3 108.78707 2.0026209 18.30954 7.108376e-35 1.937181e-32 24.61019
AMN 105.43030 1.7144345 18.26362 8.729249e-35 2.330353e-32 22.99892
CUBN 96.32447 1.3311249 18.06542 2.124577e-34 5.558319e-32 19.13330
$`Islet endocrine cells`
logFC AveExpr t P.Value adj.P.Val B
ADAMTS8 129.53805 1.5366492 26.43134 6.274147e-49 8.207212e-45 26.559205
SOST 174.42334 0.7495979 25.83500 5.271603e-48 3.447892e-44 30.636107
RIPPLY3 155.61377 2.0702438 22.83215 4.183273e-43 1.824047e-39 32.844015
SHC3 113.84060 1.3165005 22.66959 7.924827e-43 2.591616e-39 20.522624
SPARCL1 135.57671 1.4090763 22.50491 1.518573e-42 3.972891e-39 26.158014
NEFM 125.62682 3.9768428 20.09196 3.011091e-38 6.564681e-35 36.099754
ADAMTS3 105.98304 1.9854448 19.29360 9.293317e-37 1.736655e-33 19.982655
SOX2 92.95842 4.9189628 19.20445 1.369689e-36 2.239612e-33 35.061816
BMP7 65.02273 4.3748533 18.88442 5.558318e-36 8.078706e-33 26.715335
NEFL 94.09710 3.6652260 18.67045 1.428222e-35 1.868257e-32 27.924973
MUC12 82.88917 3.0025539 18.44101 3.954129e-35 4.702178e-32 21.033421
WNT1 126.77665 1.6014263 17.89197 4.646545e-34 5.065121e-31 22.254969
GDF7 128.75682 1.8171538 17.80884 6.769990e-34 6.812172e-31 23.068322
CTNNBIP1 39.15662 6.1871090 17.67897 1.221022e-33 1.140871e-30 30.833925
WNT3A 139.58409 1.2006581 17.26882 7.975502e-33 6.955170e-30 20.886580
LPL 121.11049 0.6072425 17.12012 1.583179e-32 1.294348e-29 12.929768
CBLN1 105.33229 1.2372319 16.96684 3.219255e-32 2.402381e-29 14.384192
IFIT1 88.55288 3.0191987 16.96112 3.305776e-32 2.402381e-29 20.893732
COL9A1 104.51779 2.4451146 16.24238 9.591145e-31 6.603251e-28 21.081582
LYPD1 107.78197 2.5433830 16.01103 2.874912e-30 1.880336e-27 20.834493
WNT10B 99.45440 0.8072800 15.82164 7.096417e-30 4.420392e-27 11.841066
ADGRV1 57.21112 4.3422610 15.67919 1.404254e-29 8.349569e-27 20.764235
PLEKHA5 40.69873 6.4797648 15.52835 2.900642e-29 1.649708e-26 28.721570
GAP43 75.31679 4.5145804 14.46015 5.334846e-27 2.907713e-24 23.238557
EPHA7 78.78219 3.9034653 14.43396 6.072705e-27 3.177482e-24 20.389815
TPBG 100.33057 6.1705964 14.31185 1.111856e-26 5.593916e-24 33.326828
ADCY2 95.13915 2.0573750 13.98630 5.620973e-26 2.723257e-23 11.846248
IL17RD 59.10545 3.5778488 13.90451 8.460777e-26 3.952694e-23 15.570387
UBE3D 48.01495 4.3941604 13.88658 9.255074e-26 4.060426e-23 17.585571
LMX1A 122.76479 1.1314147 13.88535 9.312192e-26 4.060426e-23 13.852762
IFITM2 52.20482 5.5045974 13.86883 1.011504e-25 4.268222e-23 22.663755
FAM181B 133.38352 3.4143865 13.54253 5.212215e-25 2.130656e-22 23.351210
SCG2 87.78095 0.7425984 13.36739 1.262241e-24 5.003447e-22 6.601143
PRCP 38.05816 5.5117030 13.35325 1.355849e-24 5.071740e-22 19.616674
CRYGS 83.79166 0.4106303 13.35308 1.357013e-24 5.071740e-22 5.603592
RBFOX1 88.75046 1.7136110 13.25294 2.253517e-24 8.188404e-22 11.716744
VAMP5 55.53149 3.9231323 13.20055 2.939515e-24 1.039238e-21 16.199904
CA10 100.98382 0.3247058 12.87761 1.520922e-23 5.235574e-21 4.861187
CDH18 99.62846 0.5513575 12.82950 1.944514e-23 6.522099e-21 7.814202
BCL2 84.60244 1.7992935 12.63277 5.320971e-23 1.740090e-20 9.677960
CD83 63.19196 2.6936773 12.62742 5.468993e-23 1.744875e-20 9.420532
IGFBP2 47.34541 8.9326234 12.49664 1.069877e-22 3.332157e-20 31.149551
FRMD3 79.98545 1.0067784 12.13875 6.758857e-22 2.056107e-19 5.380744
PCDH9 79.53995 2.6335691 12.05461 1.043919e-21 3.103525e-19 12.625904
PGAM2 59.64889 2.8311876 11.98572 1.490705e-21 4.333314e-19 9.933124
MMRN1 115.35697 1.7437715 11.96845 1.630118e-21 4.635559e-19 13.193607
MAFB 88.11635 2.8155107 11.87216 2.683885e-21 7.469767e-19 12.352118
HTRA1 68.46204 3.5987240 11.77574 4.424777e-21 1.205844e-18 13.169312
NMU 67.28790 3.1704973 11.62723 9.566725e-21 2.553925e-18 11.554305
SFRP2 59.78129 4.9878608 11.50988 1.761080e-20 4.607337e-18 17.723791
$MUC13_D
logFC AveExpr t P.Value adj.P.Val B
FAM184A 102.95228 3.7029690 20.60390 3.480193e-39 4.552440e-35 26.901403
APOA2 135.32537 4.5700960 20.28424 1.333858e-38 8.724098e-35 34.664384
FOXA2 129.82860 2.3626344 19.66000 1.906921e-37 8.314812e-34 23.070736
CST1 188.47474 3.0632741 19.00889 3.218806e-36 1.052630e-32 32.315029
CCKBR 96.54064 2.8864382 18.84157 6.711640e-36 1.755899e-32 19.794225
APOA1 160.12676 5.3603629 18.59859 1.963299e-35 4.280318e-32 38.895204
APOB 103.40119 2.4186624 18.47533 3.393978e-35 6.342374e-32 16.381441
PCBD1 55.40150 6.9252945 17.94765 3.612836e-34 5.907438e-31 31.924495
VIL1 98.62660 2.0194712 17.49520 2.823294e-33 4.103501e-30 13.119194
FOXQ1 122.03705 2.0403927 17.28702 7.334946e-33 9.594843e-30 18.133709
IHH 110.61487 1.5264822 16.92052 3.991566e-32 4.746698e-29 13.173958
EPSTI1 87.52356 3.4167152 16.88121 4.791867e-32 5.223534e-29 18.124665
PDZK1 102.13769 1.2293187 15.81284 7.401460e-30 7.447576e-27 9.554251
APOC1 73.31441 6.5875047 15.65024 1.613683e-29 1.507756e-26 28.446096
HNF4A 101.66672 1.2162260 15.57713 2.293456e-29 2.000046e-26 9.051511
PRSS2 145.60146 1.1202828 15.35733 6.624135e-29 5.415644e-26 17.077524
SERPINE2 91.48266 6.3636276 14.99486 3.857136e-28 2.967952e-25 28.554838
DENND2C 100.23173 2.2918567 14.84160 8.161992e-28 5.931501e-25 12.078375
CST4 129.04996 1.5048488 14.75038 1.276775e-27 8.790258e-25 14.445057
S100A16 83.99670 4.6850373 14.72340 1.457705e-27 9.308538e-25 20.151083
ADD3 45.34595 6.1224418 14.71834 1.494376e-27 9.308538e-25 20.703857
GATA4 90.72149 2.7684350 14.04010 4.297094e-26 2.555013e-23 13.336753
RAB25 90.26060 3.8125574 13.90390 8.486475e-26 4.826590e-23 16.426358
WFDC2 78.05954 5.5491577 13.85540 1.081916e-25 5.896892e-23 22.473366
SLC2A3 79.39011 6.2843433 13.74574 1.875111e-25 9.449104e-23 24.532227
RFX6 98.56307 0.4034583 13.74542 1.878119e-25 9.449104e-23 4.721693
KITLG 89.46728 2.8648700 13.72206 2.111827e-25 1.023141e-22 11.855039
SOX17 143.99145 2.5971138 13.64035 3.184667e-25 1.487808e-22 18.603259
ADM 85.49173 2.6525145 13.09889 4.926386e-24 2.222140e-21 10.550261
FGFR1 39.50602 6.6221206 12.93060 1.160630e-23 5.060735e-21 19.370426
FGFR4 84.70432 2.9248053 12.78172 2.482337e-23 1.047466e-20 10.940146
PRSS1 108.84084 0.8334319 12.26390 3.543666e-22 1.448584e-19 6.686932
CLDN3 73.98025 3.3835805 12.19927 4.945303e-22 1.960288e-19 10.350828
PRDM1 79.44786 2.2010891 12.06392 9.948370e-22 3.827489e-19 7.244811
MNX1 91.48632 1.5673352 12.02618 1.209190e-21 4.519260e-19 6.835304
GJA4 90.65862 1.4068779 11.97964 1.538366e-21 5.589825e-19 6.340237
FZD4 63.75631 2.5833133 11.90329 2.284222e-21 8.075649e-19 5.784391
S100A14 108.77660 2.3619882 11.86691 2.758027e-21 9.494144e-19 10.660375
CARHSP1 28.26924 7.6020921 11.85162 2.985385e-21 1.001329e-18 19.387196
DMKN 72.62431 6.0650201 11.79297 4.046430e-21 1.319979e-18 18.731492
HABP2 90.41071 0.6195381 11.78869 4.137233e-21 1.319979e-18 3.525355
PPFIBP2 58.77466 3.1999294 11.60090 1.096960e-20 3.416509e-18 7.014022
HHEX 96.14615 1.3410679 11.53735 1.526515e-20 4.587493e-18 7.294957
MGST2 56.24558 5.7152198 11.53528 1.543075e-20 4.587493e-18 15.792397
CST2 81.87065 0.8339147 11.42718 2.708386e-20 7.872976e-18 2.553418
EPCAM 95.64573 5.6005879 11.14023 1.209130e-19 3.438397e-17 17.882056
GASK1B 85.20493 1.9409450 11.02529 2.203712e-19 6.075745e-17 5.790553
LGR5 83.05251 2.0058344 11.02307 2.229461e-19 6.075745e-17 6.808390
TTN 75.05294 3.0232797 10.64108 1.644022e-18 4.388868e-16 7.549533
PLCXD3 74.53492 1.3469952 10.60624 1.973010e-18 5.161790e-16 2.526662
$`Mesangial cells`
logFC AveExpr t P.Value adj.P.Val B
TWIST1 116.82967 3.0945849 20.327052 1.113387e-38 1.456421e-34 29.730429
PRRX2 87.08075 2.7710212 18.058779 2.189079e-34 1.055745e-30 18.963311
COL6A3 116.00242 3.4290288 18.036393 2.421249e-34 1.055745e-30 28.313147
NID2 96.51343 3.8234636 16.723149 1.000967e-31 3.273412e-28 25.539942
HOXA13 109.45805 0.6432163 16.548997 2.261975e-31 5.917780e-28 11.857422
HOXA10 126.28667 0.7580840 16.500750 2.837080e-31 6.185308e-28 15.101555
ALX4 111.28615 1.0738475 16.073890 2.132089e-30 3.984265e-27 13.211826
PCOLCE 81.86994 4.8474419 15.829942 6.820231e-30 1.115193e-26 26.914598
DDR2 91.63365 2.8971187 15.330128 7.556432e-29 1.098285e-25 18.098237
EMILIN1 84.22150 2.8739884 14.479724 4.843022e-27 6.335157e-24 15.721708
BMP4 71.13083 4.6916506 13.978853 5.834093e-26 6.937798e-23 21.124887
CYB5A 55.89047 8.2444517 13.840114 1.168016e-25 1.273235e-22 32.965387
HOXA11 102.76464 0.3264158 13.206520 2.851773e-24 2.869542e-21 6.587248
PRRX1 146.73885 2.8898239 13.111484 4.620935e-24 4.317604e-21 21.687755
COL6A2 68.97292 6.2693029 12.959083 1.003759e-23 8.753445e-21 25.538797
DACT3 59.06204 3.1881491 12.591909 6.561173e-23 5.364169e-20 10.292378
VEGFC 88.91695 2.6180935 12.574949 7.157522e-23 5.507503e-20 12.824584
HOXC9 99.58343 1.0531477 12.412520 1.648615e-22 1.198085e-19 8.084623
COL1A1 79.41989 6.2764999 12.024132 1.222096e-21 8.413809e-19 23.289497
COL3A1 115.91428 5.3883411 11.874927 2.645739e-21 1.730446e-18 22.488557
HAND2 102.03943 2.7539369 11.663957 7.904901e-21 4.924000e-18 14.049291
CYB5D1 55.54760 3.8300429 11.558147 1.370037e-20 8.146117e-18 11.240163
HPSE2 80.38155 1.2954474 11.536848 1.530526e-20 8.704700e-18 5.091416
WNT5A 80.37717 3.8710975 11.515957 1.706234e-20 9.299688e-18 15.235404
ADGRA2 72.36707 2.0801881 11.506360 1.793587e-20 9.384765e-18 6.585727
S1PR3 57.49998 3.7477246 11.365801 3.728733e-20 1.875983e-17 11.074215
PCDH18 55.93164 3.4732442 11.296145 5.360730e-20 2.597174e-17 9.284439
EPB41L2 33.95835 5.7366211 11.267859 6.212652e-20 2.902418e-17 15.032487
TNS1 78.35510 1.7631603 11.207840 8.496460e-20 3.832489e-17 6.246833
KYNU 79.89166 3.0419096 11.047416 1.963194e-19 8.560180e-17 11.363501
PITX1 103.02338 4.7599224 10.991320 2.631765e-19 1.110520e-16 18.453138
COL6A1 50.05993 6.8556861 10.941577 3.413143e-19 1.395229e-16 20.459512
SERTAD4 73.45759 2.2408554 10.919701 3.826676e-19 1.516871e-16 6.919804
HOXA9 106.60821 0.5038266 10.858596 5.267254e-19 2.026498e-16 5.286805
AURKB 41.70904 5.1786100 10.738625 9.866509e-19 3.687537e-16 12.997355
KCTD12 67.44376 3.9630040 10.617705 1.858011e-18 6.751289e-16 12.109025
TBX4 78.05631 0.2957617 10.418447 5.275571e-18 1.865128e-15 1.727350
CDK6 33.91371 6.0423700 10.378490 6.504003e-18 2.238917e-15 14.668007
RARG 62.19693 2.4628392 10.179215 1.847719e-17 6.197439e-15 5.078000
SGCG 79.07164 1.1544379 10.011852 4.440856e-17 1.420810e-14 3.590516
LUM 96.09118 4.7155844 10.011319 4.453267e-17 1.420810e-14 14.972496
CTHRC1 58.06550 4.8363794 9.770576 1.571293e-16 4.893831e-14 11.947455
PGM5 82.36090 2.3608095 9.567169 4.555026e-16 1.385681e-13 7.097047
FGF20 70.04667 0.7507092 9.543386 5.158235e-16 1.533520e-13 1.245595
IGFBP4 42.33632 4.6457332 9.471204 7.522619e-16 2.151136e-13 8.809172
FREM1 71.12520 3.3016262 9.470140 7.564578e-16 2.151136e-13 8.061379
CLMP 46.65113 4.1086201 9.449748 8.415144e-16 2.342096e-13 7.893993
MIS18BP1 32.87955 5.9977414 9.412570 1.021921e-15 2.784948e-13 11.768946
IFITM2 39.45785 5.5045974 9.312745 1.721003e-15 4.594374e-13 10.822409
BAMBI 50.79116 6.5277939 9.065383 6.247927e-15 1.634583e-12 13.691003
$`Mesothelial cells`
logFC AveExpr t P.Value adj.P.Val B
SLC9A3R1 95.26541 6.6814823 38.25505 1.451018e-64 1.898076e-60 65.05152
COBLL1 109.78213 3.8078148 33.33068 1.305731e-58 8.540135e-55 43.41512
CARD16 146.36601 0.4869025 31.76006 1.477300e-56 6.441521e-53 28.79722
KRT24 151.96765 0.6689462 31.46143 3.708873e-56 1.212894e-52 30.99165
AQP1 154.68372 1.8756794 31.00990 1.511991e-55 3.955671e-52 38.02760
CD44 120.80047 2.5102088 28.87492 1.460627e-52 3.184411e-49 33.87043
FIBIN 128.17481 1.8971410 26.70966 2.351515e-49 4.394310e-46 31.21844
SLC5A12 131.74269 2.7850221 26.48915 5.113882e-49 8.361837e-46 37.10488
ANXA8 129.20971 0.8855355 25.29954 3.674710e-47 5.340987e-44 24.43782
OLR1 126.36372 0.3537423 25.02507 1.005671e-46 1.262638e-43 21.00239
SLC34A2 136.42294 0.5276665 25.01033 1.061771e-46 1.262638e-43 22.95760
TGFBR3 95.88568 3.1869881 24.19296 2.233670e-45 2.434886e-42 30.92889
SLC40A1 120.12887 3.3828801 23.96816 5.227803e-45 5.260376e-42 37.18121
MYL4 122.65111 3.2151048 22.34731 2.837965e-42 2.600522e-39 35.35263
LIMS1 49.31521 6.2651233 22.33356 2.997472e-42 2.600522e-39 40.64753
SVEP1 95.05711 2.2052858 22.31864 3.180823e-42 2.600522e-39 23.30076
ZNF641 79.94426 3.6994094 22.02784 1.016975e-41 7.825324e-39 29.69126
FRZB 131.01694 5.4381793 21.83736 2.189120e-41 1.590882e-38 47.07973
STX18 56.55477 5.5193489 21.75925 3.001450e-41 2.066419e-38 36.91805
SLC7A7 111.20631 2.5859236 21.65570 4.565913e-41 2.986336e-38 28.34319
AKAP12 80.88309 6.6390237 21.54339 7.207201e-41 4.489400e-38 47.19852
TNNT2 106.04261 2.9350416 21.33505 1.687352e-40 1.003284e-37 28.92080
TNFAIP3 87.61491 2.5601386 20.91428 9.554422e-40 5.433974e-37 22.41627
MORC4 65.81007 3.6446214 20.30462 1.223879e-38 6.670649e-36 23.84649
LEPROTL1 48.44800 6.6570553 20.04930 3.609392e-38 1.888578e-35 40.55297
GALNT10 76.85768 3.8606737 19.91599 6.368937e-38 3.204310e-35 27.99135
HSBP1L1 84.18495 3.4655492 19.86024 8.081426e-38 3.915301e-35 26.74346
DSC3 92.32285 1.9939554 19.74340 1.332818e-37 6.226640e-35 19.16457
INMT 101.81189 1.6400162 19.70742 1.555401e-37 7.015932e-35 19.09948
BCO2 90.42834 1.8742312 19.66186 1.891688e-37 8.248390e-35 17.58932
LUM 151.95763 4.7155844 19.28882 9.488361e-37 4.003782e-34 39.75903
SARAF 39.25828 7.8538281 19.12121 1.969320e-36 8.050211e-34 43.35662
JDP2 78.92256 3.6520054 19.00015 3.344447e-36 1.325719e-33 25.88144
AKR1B15 105.39495 1.5067348 18.97344 3.759846e-36 1.446546e-33 18.79866
SLN 155.68544 2.2626310 18.95779 4.027021e-36 1.505070e-33 31.23188
BNC1 104.24701 1.2420685 18.86958 5.933282e-36 2.155924e-33 17.03703
CREB3L1 90.56099 3.4812380 18.75946 9.638359e-36 3.407551e-33 26.57437
MYRF 82.88034 3.1829695 18.69162 1.300565e-35 4.393067e-33 23.04060
PXK 62.40135 3.8629424 18.69003 1.309759e-35 4.393067e-33 23.32941
IL6R 106.75300 1.5036185 18.50647 2.955136e-35 9.664033e-33 18.07065
IFIT1 92.14588 3.0191987 18.33682 6.292818e-35 2.007716e-32 23.55696
POSTN 151.33344 3.8513366 17.92884 3.933230e-34 1.225014e-31 35.00866
LRRC1 63.28836 3.6485961 17.80459 6.901817e-34 2.099597e-31 20.63788
PRSS35 98.66016 3.2397555 17.75745 8.547369e-34 2.541094e-31 25.68415
CTHRC1 85.76302 4.8363794 17.73235 9.579367e-34 2.784616e-31 32.22821
BNC2 88.35269 3.2479340 17.69253 1.147984e-33 3.264517e-31 23.68829
EGFL6 140.13891 1.8276028 17.60414 1.716882e-33 4.778412e-31 25.63787
CEBPD 91.70653 3.4107596 17.55290 2.168983e-33 5.910931e-31 25.24229
RAMP1 94.49397 3.0053905 17.52437 2.470928e-33 6.596369e-31 23.08838
DOK4 62.42553 4.8213956 17.48477 2.961329e-33 7.747429e-31 27.65548
$`Metanephric cells`
logFC AveExpr t P.Value adj.P.Val B
HOXA7 158.80141 0.44560566 22.646239 8.688901e-43 1.136595e-38 17.42554165
HOXC9 131.30059 1.05314774 17.175568 1.225620e-32 8.016166e-29 12.98481379
CDX4 136.49107 0.69810211 16.855800 5.393079e-32 2.351562e-28 11.61422218
HOXA10 133.08989 0.75808397 15.029802 3.252435e-28 1.063627e-24 10.35270820
HOXB9 157.29160 1.08866162 14.941865 4.996794e-28 1.275133e-24 15.05141625
HOXA9 139.03838 0.50382655 14.909669 5.848786e-28 1.275133e-24 9.65440425
HOXC6 130.90841 1.62292046 14.545690 3.496750e-27 6.534428e-24 13.02237384
URAD 117.96737 2.26636107 13.645606 3.101572e-25 5.071458e-22 12.66014129
HOXC8 126.24909 2.24639430 13.247679 2.314458e-24 3.363936e-21 13.43328172
HOXD9 102.76651 0.02191740 11.622086 9.825986e-21 1.285337e-17 2.22971641
HOXC4 99.01172 0.84916411 11.479458 2.063102e-20 2.453404e-17 2.89404325
HOXA5 108.66865 0.12434508 11.415118 2.883996e-20 3.143796e-17 2.44506955
HOXD4 107.02448 -0.10837864 11.295956 5.366031e-20 5.399465e-17 1.88351349
HOXC10 104.20590 0.34494651 11.158986 1.096366e-19 1.024398e-16 2.17504194
HES7 98.10440 0.61844914 10.554803 2.582814e-18 2.252386e-15 1.44466878
HOXD3 101.21432 -0.05480809 9.916242 7.327894e-17 5.991011e-14 0.28702066
HOXA6 92.09069 -0.13074454 9.756541 1.691086e-16 1.301241e-13 -0.07701277
WNT5A 80.79595 3.87109754 9.359924 1.345311e-15 9.776677e-13 7.58732351
FZD10 92.66475 1.64816950 9.244160 2.461415e-15 1.694620e-12 2.27484052
CDX2 110.22885 3.39599403 9.193238 3.209945e-15 2.099464e-12 8.13425440
SALL1 66.83448 3.52940145 9.149307 4.035774e-15 2.513903e-12 4.55843812
FGF8 90.74090 2.27163976 8.844193 1.972431e-14 1.172790e-11 3.22511693
BMP4 60.29008 4.69165060 8.450799 1.508590e-13 8.374926e-11 5.22620368
HOXB3 93.07565 2.81409534 8.447232 1.536566e-13 8.374926e-11 4.31763909
FOXF2 79.38915 1.72055207 8.300967 3.260538e-13 1.706044e-10 1.15642660
FOXB1 81.75363 1.17327676 8.243733 4.373682e-13 2.200467e-10 -0.64773876
HOXD8 87.23346 -0.07212554 8.108583 8.736540e-13 4.232692e-10 -1.28104130
MLLT3 62.22285 5.07183549 7.963603 1.830183e-12 8.550221e-10 5.77002038
RARG 64.28507 2.46283917 7.851474 3.235643e-12 1.459498e-09 -0.13842061
PRRX2 61.17551 2.77102125 7.825529 3.690618e-12 1.609232e-09 0.51598081
HOXA3 108.15130 0.54333794 7.804023 4.115547e-12 1.736628e-09 0.41333637
FGF17 120.97491 2.12486885 7.754855 5.278511e-12 2.157756e-09 3.99247093
HOXB8 99.36657 2.17965111 7.725072 6.136143e-12 2.432330e-09 2.54130507
MCM7 34.39258 6.55663259 7.606184 1.117501e-11 4.299419e-09 5.78407068
HOXB7 84.81812 2.06936902 7.506908 1.839972e-11 6.876763e-09 0.84533700
GTF3A 23.25751 7.08828325 7.446532 2.489581e-11 9.046168e-09 5.86878932
MCM3 28.34464 6.37609094 7.371435 3.622700e-11 1.280771e-08 4.57232979
PRRX1 118.25370 2.88982390 7.323080 4.609643e-11 1.586809e-08 3.96796613
NASP 23.94041 8.81893924 7.309735 4.926142e-11 1.652278e-08 8.33787422
MCM4 33.76286 5.61462224 7.290575 5.418615e-11 1.772023e-08 3.22453738
TGFBI 74.10172 3.29600219 7.169415 9.880399e-11 3.152329e-08 1.23164197
HNRNPD 16.04178 8.77013265 7.082118 1.520129e-10 4.734477e-08 7.15132276
HOXA4 77.91954 -0.04234780 7.051897 1.763923e-10 5.366018e-08 -2.03819855
HOXA11 80.77753 0.32641585 6.971940 2.611820e-10 7.764823e-08 -1.81744140
SKAP2 60.37785 1.98994353 6.955780 2.826936e-10 8.217590e-08 -1.14420591
NID2 63.93392 3.82346364 6.920700 3.356095e-10 9.543713e-08 1.66037591
CENPF 46.35478 8.07651079 6.770823 6.961661e-10 1.937564e-07 6.90678004
SALL4 41.47067 5.74912108 6.729399 8.508477e-10 2.318737e-07 3.22378683
MSGN1 91.22716 0.43441408 6.675921 1.101673e-09 2.941016e-07 -1.80709977
DEK 21.03932 8.03922184 6.650019 1.248178e-09 3.265484e-07 5.20493264
$`Retinal pigment cells`
logFC AveExpr t P.Value adj.P.Val B
SFRP2 141.32830 4.98786079 36.33907 2.492096e-62 3.259911e-58 58.246252
DCT 183.61646 0.92656067 26.51734 4.629126e-49 2.303410e-45 32.776867
CCL2 136.54778 2.53603706 26.47996 5.282647e-49 2.303410e-45 34.919564
RAX 155.02632 0.81413097 26.00803 2.832239e-48 9.262130e-45 27.200686
DAPL1 169.08091 0.64783606 24.57243 5.382477e-46 1.408164e-42 28.013147
SIX6 163.56354 0.85100235 22.86087 3.737934e-43 8.149318e-40 26.992171
MITF 108.93119 2.41605164 22.72373 6.403785e-43 1.196684e-39 26.329393
CLEC19A 119.34980 0.36711015 21.81131 2.431918e-41 3.976489e-38 16.678809
HOMER2 86.08452 2.81496580 21.50547 8.410835e-41 1.222468e-37 23.523422
ALDH1A1 142.61414 2.18517354 19.87570 7.564462e-38 9.895072e-35 28.225173
PMEL 139.53463 4.87775775 19.32253 8.195613e-37 9.746074e-34 41.143281
CCKBR 89.26744 2.88643816 18.87949 5.680199e-36 6.191890e-33 22.492708
DBX2 112.03552 -0.01289448 18.29197 7.689521e-35 7.737433e-32 10.700173
PGAM2 74.09051 2.83118756 16.26100 8.782972e-31 8.206433e-28 16.640217
AL391650.1 109.10959 1.17877935 15.64053 1.690746e-29 1.474443e-26 14.816826
SYNPR 97.70663 0.48089549 14.93305 5.216913e-28 4.265152e-25 8.737079
MAB21L1 102.73132 1.23913399 14.66734 1.920325e-27 1.477634e-24 12.139598
TRPM3 102.55519 2.49547548 14.46031 5.330624e-27 3.873883e-24 18.196964
SIX3 122.14977 2.60080335 14.16696 2.283704e-26 1.572270e-23 19.577923
RAB27A 65.41879 2.73489754 14.13558 2.669800e-26 1.746182e-23 10.697499
TYR 95.96543 -0.08943723 13.97099 6.067934e-26 3.779745e-23 6.701123
TYRP1 106.50732 1.41698662 13.34262 1.430773e-24 8.290311e-22 12.718529
SERPINF1 78.13441 5.73659766 13.33726 1.470131e-24 8.290311e-22 25.846696
SOX2 72.25486 4.91896284 13.33054 1.521042e-24 8.290311e-22 21.653396
PLCH1 66.94320 2.87856906 13.31750 1.624793e-24 8.501565e-22 12.749677
ATP6V1C2 96.37624 1.19716966 13.26123 2.160806e-24 1.087135e-21 11.980023
SPOCK1 76.22616 3.06394163 12.77136 2.617349e-23 1.268057e-20 13.681686
BMP7 50.76819 4.37485330 12.72694 3.285079e-23 1.534718e-20 14.596731
DIPK1C 113.66888 0.46823211 12.30052 2.934204e-22 1.303677e-19 8.728931
CRABP1 106.65448 6.11607830 12.29688 2.989856e-22 1.303677e-19 26.831918
MYO3B 73.90201 1.03301423 12.15827 6.110711e-22 2.578523e-19 5.766586
SFRP1 66.19226 6.27492262 12.11178 7.769063e-22 3.175847e-19 23.253401
HCRT 81.20656 0.64094386 12.00438 1.353542e-21 5.365360e-19 5.743734
EFNA5 72.22309 5.18781468 11.91943 2.100975e-21 8.083192e-19 19.952141
CLDN1 72.68037 1.53908082 11.82592 3.410856e-21 1.274783e-18 7.727196
ANXA1 115.38067 3.63810634 11.65727 8.184363e-21 2.973879e-18 17.782423
CRX 83.21879 0.28441292 11.52890 1.595107e-20 5.639350e-18 3.700153
SNCA 55.07566 4.74965867 11.49838 1.869634e-20 6.391661e-18 15.564666
TSPAN10 69.65780 2.46668310 11.49472 1.905625e-20 6.391661e-18 9.926649
LAMP5 83.57894 0.69345385 11.43581 2.589448e-20 8.468141e-18 3.786323
MAB21L2 78.80199 2.77646879 11.41038 2.956109e-20 9.431429e-18 11.288639
EFEMP1 68.29351 2.13398293 11.37250 3.600804e-20 1.121479e-17 6.770419
STRIP2 61.82783 1.70241747 11.15073 1.144671e-19 3.482196e-17 5.255785
ALDH1A3 104.69322 1.01266988 10.94704 3.317025e-19 9.861364e-17 6.784592
TRPM1 76.92175 -0.26571923 10.90268 4.182929e-19 1.215931e-16 2.578834
PAX6 111.96694 3.52118809 10.86096 5.202587e-19 1.479458e-16 15.750825
TCEAL7 68.18248 4.22573100 10.83887 5.839717e-19 1.600459e-16 13.246197
S100A11 -63.50749 10.01712682 -10.83779 5.872795e-19 1.600459e-16 16.263682
PTPRZ1 73.73152 3.76794437 10.82928 6.140223e-19 1.639189e-16 12.713017
SEMA3A 53.66653 4.37055799 10.81793 6.515743e-19 1.704649e-16 12.831813
$`SLC24A4_PEX5L positive cells`
logFC AveExpr t P.Value adj.P.Val B
GNG3 152.37451 2.6673015 45.39624 4.087886e-72 5.347364e-68 40.71307
INA 149.62034 2.6686304 40.07271 1.346128e-66 8.804353e-63 39.55716
STMN4 182.63416 1.8398121 36.62746 1.131932e-62 4.935601e-59 39.56391
TUBB2B 90.03668 8.1127821 34.86748 1.521483e-60 4.975630e-57 67.76138
TMEM163 148.20156 1.1117885 33.73003 4.040031e-59 1.056953e-55 26.56300
SLC17A6 156.20164 0.7775274 33.60118 5.891399e-59 1.284423e-55 26.52526
RTN1 162.71114 3.1365014 32.33098 2.591724e-57 4.843192e-54 42.78340
CRMP1 131.18170 3.7555245 32.15584 4.408636e-57 7.208671e-54 40.43336
NSG2 159.14008 0.4217989 31.09659 1.152982e-55 1.675795e-52 24.35030
KIF5C 125.61170 4.4306399 30.62587 5.060995e-55 6.620287e-52 43.83786
CDKN2D 100.41004 3.4783630 30.55754 6.282688e-55 7.471258e-52 30.94207
GAP43 136.48913 4.5145804 30.50016 7.535670e-55 8.214509e-52 46.45598
PCSK1N 136.60869 3.7471102 30.16376 2.200818e-54 2.214530e-51 40.56394
DCX 148.85898 2.6276693 30.10285 2.674899e-54 2.499311e-51 36.24807
OLFM1 132.03483 2.7883832 29.71992 9.183655e-54 8.008759e-51 33.78192
TLCD3B 140.62775 1.7731339 29.65057 1.149783e-53 9.400194e-51 27.47940
ELAVL3 145.53091 2.1343713 29.51288 1.798541e-53 1.383925e-50 31.67273
TTC9B 154.83111 0.4408217 29.32343 3.337560e-53 2.425479e-50 22.03152
CELF3 141.56814 1.2546600 29.18507 5.252814e-53 3.616425e-50 24.98545
UNCX 150.78647 0.8871815 28.60949 3.528499e-52 2.307815e-49 24.08694
SYP 137.22088 1.8687858 27.96024 3.134645e-51 1.952585e-48 27.29609
LHX9 151.27418 2.0780892 27.66748 8.499853e-51 4.886776e-48 31.62018
STMN2 170.33950 4.7054494 27.66432 8.592299e-51 4.886776e-48 53.31682
MAP1B 87.32345 8.7464887 26.84085 1.484573e-49 8.091542e-47 64.42512
MAB21L1 152.69616 1.2391340 26.68745 2.542387e-49 1.330279e-46 26.26312
MLLT11 90.24958 7.5497265 26.37986 7.529071e-49 3.787991e-46 56.64743
KLC1 56.92669 5.7901257 26.32269 9.221996e-49 4.467886e-46 35.60447
MYT1L 141.83040 0.4248931 25.28321 3.900742e-47 1.822343e-44 18.13178
PCSK2 138.60491 1.2535270 25.22275 4.866509e-47 2.195131e-44 22.25312
ADCYAP1 133.25161 0.8113424 25.18616 5.564609e-47 2.426355e-44 18.90266
CDK5R2 121.62164 0.4570845 23.88474 7.177352e-45 3.028611e-42 12.69251
INSM1 119.44623 1.9175950 23.70350 1.432730e-44 5.856731e-42 21.67223
GNAO1 113.22976 1.3581541 23.66724 1.645894e-44 6.524224e-42 16.49048
SNAP25 113.30137 1.6462144 22.67535 7.747204e-43 2.980623e-40 18.00343
GDAP1L1 139.72309 1.4553746 22.40688 2.239788e-42 8.371048e-40 22.51823
POU2F2 105.77498 2.4044080 22.25935 4.028113e-42 1.463660e-39 19.76063
NEFL 113.34640 3.6652260 22.08429 8.109424e-42 2.867010e-39 30.55475
GDAP1 82.54007 3.3711802 21.93249 1.491913e-41 5.135713e-39 20.72740
TAGLN3 140.61725 3.8492674 21.78076 2.751485e-41 9.152310e-39 36.41540
PRPH 122.85085 2.2754711 21.77655 2.798658e-41 9.152310e-39 24.64320
NRXN1 122.97923 2.7210766 21.62150 5.245896e-41 1.673697e-38 26.01677
NDRG4 103.59407 2.6253264 21.57340 6.378749e-41 1.986677e-38 20.28409
NSG1 100.33655 4.1692336 21.35639 1.546149e-40 4.703529e-38 30.21726
NHLH2 109.85331 1.9647414 21.23824 2.509788e-40 7.461485e-38 18.42603
ELAVL2 114.95598 2.7520644 21.18410 3.135266e-40 9.113869e-38 24.59542
SCN3B 111.49643 0.9519570 21.10589 4.326750e-40 1.230396e-37 12.47056
TUBA1A 93.41180 9.5832524 21.08310 4.753234e-40 1.322916e-37 59.67462
SYT1 108.41512 4.4850794 20.96972 7.593853e-40 2.067499e-37 33.90290
KLHL35 118.94251 2.1556421 20.96497 7.744625e-40 2.067499e-37 21.19815
ACTL6B 127.67692 0.6081244 20.83884 1.306778e-39 3.418793e-37 13.89705
$`Squamous epithelial cells`
logFC AveExpr t P.Value adj.P.Val B
KRT7 176.08989 0.74277684 33.07127 2.815132e-58 3.682474e-54 31.589294
TACSTD2 179.76022 1.12840782 31.91331 9.236358e-57 6.041040e-53 33.530498
SFN 157.47289 1.34530658 30.16803 2.170976e-54 9.466179e-51 30.077340
TINCR 142.00712 1.03798232 30.07518 2.923049e-54 9.559102e-51 23.745486
NECTIN4 142.03122 0.56824527 29.51662 1.776755e-53 4.648347e-50 22.712738
WNT6 173.59437 0.50925547 26.86826 1.348842e-49 2.940700e-46 28.032573
GABRP 160.32536 0.57486256 25.92246 3.849477e-48 7.193573e-45 24.999699
ITGB6 142.93595 0.23176538 25.69836 8.628281e-48 1.410832e-44 18.071851
CLDN4 155.16655 2.92753410 25.64577 1.043533e-47 1.516717e-44 36.564768
LYPD3 143.36843 0.70675295 25.55490 1.450368e-47 1.897227e-44 21.474028
VGLL1 144.69168 0.09355883 25.30054 3.661278e-47 4.353925e-44 17.880676
VTCN1 158.83857 0.34406005 25.17310 5.837468e-47 6.363326e-44 23.370408
TMEM40 135.77703 0.15551975 23.65300 1.738156e-44 1.748986e-41 14.330060
MUC15 139.28388 0.11722375 23.47131 3.491804e-44 3.262592e-41 18.141643
GPRC5A 141.22647 0.40084966 22.01680 1.063067e-41 9.270649e-39 15.657218
TMEM54 90.53047 3.58307930 21.72509 3.446451e-41 2.817689e-38 26.030953
EPAS1 108.40412 2.45353489 21.68355 4.078285e-41 3.138121e-38 22.968469
RAB25 113.42922 3.81255735 21.27318 2.174404e-40 1.580187e-37 31.255957
S100A16 101.93139 4.68503728 21.00140 6.661194e-40 4.586056e-37 33.485563
TPM1 68.62426 8.87944070 20.16687 2.191434e-38 1.433308e-35 51.013640
S100A11 63.49027 10.01712682 20.10143 2.892381e-38 1.801678e-35 54.961971
KRT18 89.25220 8.52937448 19.87674 7.531061e-38 4.477901e-35 48.951743
PRKCH 98.71404 0.99175495 19.68470 1.714830e-37 9.752910e-35 9.983866
ERP27 114.33751 1.31781648 19.58426 2.641856e-37 1.439922e-34 15.983466
TRIM55 115.43699 0.37550628 19.42297 5.302435e-37 2.774446e-34 11.000128
KRT8 74.31422 8.88754755 19.12174 1.964722e-36 9.884821e-34 48.493521
RHOV 116.14032 1.20764426 18.89953 5.201112e-36 2.519843e-33 14.384246
ZNF750 118.74542 0.08346852 18.84414 6.636192e-36 3.100287e-33 10.701398
CLIC3 128.43552 1.68014670 18.64663 1.586965e-35 7.158307e-33 19.417281
EPN3 111.69865 0.39052705 18.59390 2.004574e-35 8.740610e-33 9.382514
XAGE2 135.31802 0.25657422 18.54110 2.533748e-35 1.069160e-32 12.621753
SPNS2 99.70094 0.89107234 18.34662 6.023484e-35 2.443028e-32 10.692497
CST6 151.47402 0.76838894 18.34148 6.163131e-35 2.443028e-32 17.720707
SMAGP 76.67694 3.49492966 18.28699 7.862563e-35 3.025005e-32 19.538724
KRT19 76.57681 8.67978966 18.23837 9.773872e-35 3.652915e-32 44.790067
ELF3 97.77304 3.23883569 17.99717 2.889365e-34 1.049883e-31 22.047183
ANXA3 102.86632 4.34168451 17.92027 4.088549e-34 1.445468e-31 28.149456
TINAGL1 120.46821 0.81955876 17.75960 8.464448e-34 2.913775e-31 12.610202
CYP1A1 116.64446 0.68297805 17.73517 9.457487e-34 3.172138e-31 12.444150
P2RY6 114.57426 1.22280370 17.70320 1.093624e-33 3.576423e-31 13.385563
ST14 116.28871 1.53593571 17.62176 1.584406e-33 4.994180e-31 14.925605
PLAU 106.13067 1.91522027 17.61913 1.603513e-33 4.994180e-31 16.203624
S100A9 168.44350 0.31810941 17.53447 2.359535e-33 7.177925e-31 18.927392
ANXA1 157.48290 3.63810634 17.50373 2.715334e-33 8.072564e-31 31.249388
TACC1 74.97340 3.71678785 17.37375 4.924529e-33 1.431506e-30 19.419911
CLDN3 88.72676 3.38358047 17.36078 5.226465e-33 1.486247e-30 20.401627
ELF5 112.43417 -0.11482346 17.22766 9.639814e-33 2.682945e-30 7.927264
ARHGAP29 70.19983 5.06595765 17.19085 1.142208e-32 3.112755e-30 26.422630
EGFR 82.92879 2.11760612 17.08140 1.893568e-32 5.055053e-30 11.263889
PTGES 111.41585 1.41220432 16.91180 4.156591e-32 1.087447e-29 13.512721
$`Stellate cells`
logFC AveExpr t P.Value adj.P.Val B
ANGPTL6 125.67056 1.2356458 30.14263 2.354817e-54 3.080337e-50 27.81265
SLC9A3R1 69.49483 6.6814823 25.39139 2.628241e-47 1.719001e-43 50.05698
COLEC10 120.01790 0.3153389 24.14257 2.701383e-45 1.177893e-41 17.89767
TGFBR3 94.40351 3.1869881 24.06400 3.635654e-45 1.188950e-41 31.28339
COLEC11 115.33599 1.6066477 23.88001 7.307783e-45 1.911862e-41 24.73567
FIBIN 117.75717 1.8971410 23.60887 2.058413e-44 4.487684e-41 27.82765
COBLL1 82.41871 3.8078148 22.42258 2.104447e-42 3.932609e-39 31.65674
SVEP1 90.95072 2.2052858 20.95363 8.117182e-40 1.194500e-36 21.77151
CPED1 119.99761 2.0788448 20.95063 8.218408e-40 1.194500e-36 27.41142
TSLP 123.91560 1.6183551 20.77620 1.695613e-39 2.218032e-36 25.15464
LUM 157.93497 4.7155844 20.50950 5.168471e-39 6.146252e-36 42.56861
DOK4 68.59864 4.8213956 20.20463 1.867550e-38 2.035785e-35 32.86736
FOXF1 108.38694 1.7924233 19.97535 4.944579e-38 4.975388e-35 22.61515
KCNE4 109.04846 0.5312068 19.83291 9.083128e-38 8.486885e-35 14.64908
COL1A1 111.23057 6.2764999 19.60776 2.387555e-37 2.082107e-34 44.72722
TEK 112.48944 1.4671969 19.34306 7.496899e-37 6.129184e-34 20.67891
JDP2 77.32241 3.6520054 18.75651 9.764376e-36 7.513400e-33 25.90236
ADGRA2 91.19337 2.0801881 18.68495 1.339494e-35 9.734399e-33 18.91592
CYB5D1 72.78877 3.8300429 18.60988 1.867475e-35 1.285707e-32 25.77001
CTHRC1 87.92594 4.8363794 18.44805 3.832214e-35 2.506459e-32 33.83901
ZNF641 70.05060 3.6994094 18.39708 4.809054e-35 2.995583e-32 24.57582
AKAP12 70.79968 6.6390237 18.23995 9.704788e-35 5.770379e-32 40.24017
LDB2 91.42219 3.3761267 18.13909 1.525621e-34 8.676803e-32 25.84461
IGF2 136.40483 3.4009741 18.02620 2.534987e-34 1.381673e-31 32.26641
PAMR1 83.71323 2.9790698 17.95744 3.456695e-34 1.808681e-31 22.27806
PRDM6 91.47760 1.8387960 17.87196 5.086633e-34 2.559163e-31 17.32527
CEBPD 91.54645 3.4107596 17.77883 7.757076e-34 3.758160e-31 26.00752
DSC3 85.02992 1.9939554 17.58263 1.893786e-33 8.847363e-31 16.57943
NID2 96.15464 3.8234636 17.51582 2.569390e-33 1.158972e-30 28.30090
ATP1B1 47.03459 7.4211313 17.50683 2.677139e-33 1.167322e-30 39.35622
PROM1 72.25689 3.8433103 17.45551 3.385563e-33 1.428598e-30 23.92344
CD248 91.69274 2.2968251 17.43746 3.677205e-33 1.503172e-30 19.37771
MMP1 115.90186 1.3810578 17.38973 4.576444e-33 1.814074e-30 19.03211
CREB3L1 85.06153 3.4812380 17.34846 5.530669e-33 2.127844e-30 24.44471
POSTN 146.69901 3.8513366 17.29813 6.969722e-33 2.604884e-30 33.77001
COL6A3 108.89155 3.4290288 17.18389 1.179448e-32 4.285655e-30 27.68771
BMP4 78.36677 4.6916506 17.09123 1.809374e-32 6.396872e-30 29.29876
DDR2 92.73230 2.8971187 16.82709 6.164394e-32 2.122011e-29 22.27428
SLC40A1 93.69839 3.3828801 16.81543 6.508444e-32 2.182999e-29 25.38491
LRRC32 86.12294 2.2407753 16.73722 9.373010e-32 3.065208e-29 17.33403
AFAP1L2 86.95883 2.6132810 16.51049 2.710205e-31 8.646877e-29 19.15816
KCTD12 87.44824 3.9630040 16.48776 3.015731e-31 9.392566e-29 26.52097
PSD 76.26699 2.9797715 16.37865 5.039793e-31 1.533152e-28 19.04034
COL1A2 91.17029 6.9908439 16.34674 5.858252e-31 1.741632e-28 40.40106
RGS1 102.52738 1.1881097 16.30337 7.189108e-31 2.089794e-28 14.47996
SYT10 95.77620 2.6471171 16.29523 7.470910e-31 2.124499e-28 20.56872
DACT3 65.97882 3.1881491 16.28339 7.900815e-31 2.198948e-28 17.81436
IGFBP3 118.92160 4.8257654 16.19753 1.186010e-30 3.232123e-28 32.89110
HAND1 134.77807 4.6368572 16.13150 1.621890e-30 4.329785e-28 32.94707
SIPA1L2 58.33261 6.1730156 16.11794 1.729646e-30 4.525099e-28 32.74235
$`Stromal cells`
logFC AveExpr t P.Value adj.P.Val B
TNNT2 108.18348 2.9350416 20.58525 3.762754e-39 4.922058e-35 26.142266
COL1A1 117.06049 6.2764999 19.65980 1.908557e-37 1.248292e-33 43.009352
DDR2 107.77230 2.8971187 19.53850 3.218130e-37 1.403212e-33 25.026258
COBLL1 79.47168 3.8078148 19.32050 8.268170e-37 2.703898e-33 24.610660
COL6A3 123.21770 3.4290288 19.09778 2.181581e-36 5.707452e-33 29.484778
CYB5D1 78.79138 3.8300429 18.95405 4.093636e-36 8.924809e-33 23.971441
SLC9A3R1 55.63827 6.6814823 17.53011 2.406958e-33 4.497917e-30 33.306938
DSC3 89.69937 1.9939554 17.25547 8.481054e-33 1.386758e-29 14.313306
PCOLCE 89.36123 4.8474419 17.21487 1.022486e-32 1.486126e-29 28.955816
BMP4 82.04699 4.6916506 16.78545 7.484385e-32 9.790324e-29 26.716740
LUM 139.97848 4.7155844 16.35845 5.543392e-31 6.592101e-28 31.772626
RGS4 116.93518 3.5193512 16.19133 1.221353e-30 1.331377e-27 26.070572
NID2 95.50658 3.8234636 16.03810 2.527523e-30 2.543271e-27 23.599679
COL5A1 85.56052 4.4965267 15.71006 1.210970e-29 1.131479e-26 25.129675
COL3A1 143.18072 5.3883411 15.50759 3.205969e-29 2.795818e-26 32.648309
POSTN 138.85322 3.8513366 14.95951 4.583870e-28 3.747600e-25 26.684269
FGFR2 57.27743 4.9602560 14.78903 1.056183e-27 8.127021e-25 20.827042
COL6A2 78.70748 6.2693029 14.67909 1.812513e-27 1.317194e-24 29.383711
RGS13 136.54020 2.6808708 14.54611 3.489540e-27 2.289248e-24 21.823553
DOK4 58.14475 4.8213956 14.54550 3.500112e-27 2.289248e-24 20.020293
BNC2 80.51735 3.2479340 14.39787 7.260020e-27 4.522301e-24 16.469757
CHN1 60.06911 5.1116585 14.30461 1.152506e-26 6.852694e-24 21.899846
TNNI1 112.23626 3.9575031 13.93017 7.441303e-26 4.232160e-23 22.835920
MFAP4 96.90398 4.0818825 13.87363 9.874933e-26 5.382250e-23 21.712842
TCF21 92.63423 1.8131395 13.82104 1.285212e-25 6.724743e-23 11.169983
ADGRA2 80.55678 2.0801881 13.80508 1.392240e-25 7.004573e-23 10.166371
HAND1 125.69577 4.6368572 13.77658 1.606174e-25 7.781618e-23 25.511846
COL1A2 85.35443 6.9908439 13.75237 1.813669e-25 8.473075e-23 30.829088
DCN 143.40029 3.1437291 13.63822 3.218980e-25 1.451982e-22 22.038099
CYB5A 57.22075 8.2444517 13.61901 3.545817e-25 1.546094e-22 31.435052
CRISPLD2 82.53344 1.9406762 13.57082 4.519746e-25 1.907187e-22 9.667454
CLEC1B 116.42553 0.9545363 13.33793 1.465142e-24 5.989227e-22 10.652023
RPP25 57.71231 4.6152140 13.27267 2.039045e-24 8.082651e-22 17.309567
KCTD12 79.83735 3.9630040 13.20262 2.908709e-24 1.119083e-21 17.672909
CREB3L1 75.63196 3.4812380 13.19183 3.072522e-24 1.148333e-21 14.676387
COL21A1 90.22203 2.4613167 13.16725 3.480838e-24 1.264801e-21 12.524578
PRDM6 79.98501 1.8387960 13.04877 6.357081e-24 2.247486e-21 8.804129
EMILIN1 80.90369 2.8739884 13.04257 6.560920e-24 2.258510e-21 12.459458
HAND2 110.55522 2.7539369 12.98581 8.759329e-24 2.937969e-21 16.871432
AKR1B15 89.95620 1.5067348 12.72707 3.282894e-23 1.073588e-20 7.769805
ATP7B 66.83761 3.0250264 12.68044 4.167975e-23 1.305883e-20 10.326147
PITX1 116.93007 4.7599224 12.67928 4.192883e-23 1.305883e-20 22.903836
AKAP4 88.88907 0.7453011 12.62267 5.603748e-23 1.704712e-20 6.021759
TWIST1 86.62156 3.0945849 12.57185 7.272234e-23 2.162002e-20 14.521911
BAG2 51.54023 4.7561435 12.41755 1.606487e-22 4.669880e-20 15.427152
RGS1 91.25725 1.1881097 12.40382 1.724106e-22 4.902832e-20 7.590996
ACTA2 91.09013 4.4290611 12.22057 4.430902e-22 1.233205e-19 18.733755
GPX8 39.23205 6.1566286 12.14119 6.674252e-22 1.813202e-19 18.927450
COL6A1 56.65777 6.8556861 12.13780 6.792057e-22 1.813202e-19 23.331080
TMEM88 94.80654 6.3716813 12.03416 1.160328e-21 3.035651e-19 24.178627
$`Thymic epithelial cells`
logFC AveExpr t P.Value adj.P.Val B
CER1 193.00176 2.03976580 30.695676 4.059492e-55 5.310221e-51 36.5694701
GYPB 177.99350 0.42110411 25.977722 3.157169e-48 2.064946e-44 23.6216149
APOBEC3G 114.37166 2.09624580 23.010184 2.085013e-43 9.091350e-40 17.9371052
MIXL1 171.00686 2.28064846 22.653002 8.460322e-43 2.766737e-39 29.6043258
PLSCR2 175.71224 1.21188125 22.243069 4.298355e-42 1.124536e-38 25.2961767
SHISAL2B 116.01167 2.60023393 22.043843 9.537185e-42 2.079265e-38 20.6421597
MESP1 145.98008 2.43703522 19.555526 2.990275e-37 5.587969e-34 24.3232941
NTS 147.06861 2.64617511 19.055155 2.628553e-36 4.298012e-33 24.5873159
CYP26A1 147.71944 1.52578314 18.586385 2.072514e-35 3.012285e-32 19.0191666
MESP2 124.06967 1.26059252 17.005968 2.685037e-32 3.512297e-29 12.0882585
GYPE 124.69905 0.55809555 16.261085 8.779308e-31 1.044019e-27 7.9246891
SERPINE2 95.63232 6.36362760 15.135771 1.940941e-28 2.115787e-25 27.8114689
APLNR 133.57700 2.34443547 14.958026 4.617334e-28 4.646104e-25 16.9070604
HS3ST3A1 91.30279 2.11167551 14.884777 6.606422e-28 6.172758e-25 8.6556701
CD48 112.10588 0.33866121 14.663329 1.958592e-27 1.708023e-24 5.3706039
P3H2 77.71862 3.73554197 14.425083 6.345189e-27 5.187588e-24 13.2798296
TRDN 121.88235 2.43023177 14.014042 4.893833e-26 3.765660e-23 14.3395342
SERHL2 96.09632 1.01740361 13.382580 1.168904e-24 8.494684e-22 3.9673889
ARL4D 66.74664 5.51507338 13.226595 2.575631e-24 1.773254e-21 17.9992208
GYPA 107.98440 -0.03156862 12.706006 3.656604e-23 2.391602e-20 2.1105850
EOMES 135.43803 2.44980940 12.472010 1.214226e-22 7.563469e-20 14.3365573
GSC 115.73073 1.21157204 12.360318 2.156584e-22 1.282285e-19 7.0830196
FOXH1 108.41923 3.99993072 12.179629 5.472914e-22 3.112660e-19 15.5705632
CD1D 101.50916 1.51693622 12.139658 6.727143e-22 3.666573e-19 5.1843372
LZTS1 84.78803 2.19411719 12.091050 8.647134e-22 4.524527e-19 5.6812250
GRP 112.31277 0.83584052 11.801895 3.863369e-21 1.943721e-18 4.3560725
FGF17 149.07288 2.12486885 11.692900 6.801665e-21 3.295281e-18 13.7242619
CCDC81 89.03593 2.00758838 11.604642 1.075845e-20 5.026116e-18 5.1481383
FAM24B 114.43025 0.88054893 11.483357 2.021665e-20 9.119105e-18 4.8812022
BMP4 71.13815 4.69165060 11.282778 5.747682e-20 2.506181e-17 11.6999690
TBX6 92.46735 0.91899072 11.273858 6.021319e-20 2.540802e-17 1.6637827
CCKBR 76.72863 2.88643816 11.263807 6.345326e-20 2.593850e-17 6.2252316
ZIC3 75.29811 3.58296979 11.239344 7.208795e-20 2.857523e-17 7.9610729
NR0B1 88.14092 0.75819062 11.207892 8.494137e-20 3.267994e-17 1.5431449
JAKMIP1 88.02260 0.84945015 11.129435 1.279229e-19 4.649940e-17 1.6689264
RFPL2 99.71961 0.13586254 11.129364 1.279702e-19 4.649940e-17 0.9251215
TESC 86.15826 2.45226621 10.872931 4.886840e-19 1.727696e-16 5.2296171
GAL 96.90344 4.49434675 10.750882 9.253526e-19 3.185405e-16 13.6467835
NFKBIA 41.00109 5.55770686 10.562961 2.474797e-18 8.300723e-16 9.7357296
LRRTM1 87.47936 1.16791958 10.486523 3.693181e-18 1.207763e-15 1.7498090
DLL3 59.48401 4.30887700 10.449541 4.482605e-18 1.430170e-15 8.0782715
TMOD1 80.77850 1.92937139 10.239819 1.345007e-17 4.189056e-15 2.8636541
APCDD1L 87.07631 0.73781400 10.013539 4.401784e-17 1.339064e-14 0.4367276
LAPTM4B 33.74561 8.86686851 9.976650 5.340181e-17 1.587611e-14 16.9758623
TUBB2B 37.25719 8.11278211 9.918652 7.235995e-17 2.103423e-14 15.4807605
CKM 87.43050 1.17161988 9.883336 8.706243e-17 2.475791e-14 1.2437795
GATA6 84.79394 4.19882434 9.866547 9.506381e-17 2.645808e-14 9.5972553
HAS2 75.48560 5.59983355 9.795489 1.379148e-16 3.743434e-14 11.9942027
ACSS3 59.09036 3.23960505 9.792316 1.402250e-16 3.743434e-14 3.3384154
VRTN 96.43963 2.17742682 9.734739 1.895526e-16 4.959075e-14 4.7000619
$`Visceral neurons`
logFC AveExpr t P.Value adj.P.Val B
INSM1 143.48626 1.9175950 29.21276 4.796400e-53 6.274171e-49 24.744458
PPP1R17 184.54560 1.8014410 28.93940 1.180041e-52 7.718058e-49 32.844050
GNG3 123.50502 2.6673015 28.45939 5.826723e-52 2.540645e-48 24.352301
ELAVL3 149.72484 2.1343713 27.66509 8.569664e-51 2.802494e-47 26.827052
STMN4 162.39350 1.8398121 27.42875 1.928683e-50 5.045822e-47 28.068321
TLX3 166.65386 0.7263713 26.64117 2.991755e-49 6.522524e-46 22.208885
NHLH1 171.36370 2.4191008 26.08902 2.119770e-48 3.961244e-45 32.121334
SST 213.42537 2.5685871 25.37987 2.740948e-47 4.481793e-44 39.711203
SNCG 169.17925 3.0455669 23.95076 5.584592e-45 8.116895e-42 33.710776
POU4F1 164.98584 1.1963388 23.69237 1.495019e-44 1.955635e-41 22.734270
TLX2 145.10099 0.6052476 23.64412 1.798289e-44 2.138493e-41 14.660848
STMN2 163.01898 4.7054494 23.25301 8.112485e-44 8.843284e-41 42.571324
ELAVL2 130.15549 2.7520644 23.13255 1.294695e-43 1.302762e-40 24.471039
SIX1 167.59031 1.5451431 23.08783 1.540662e-43 1.439528e-40 25.519121
THSD7B 140.24223 0.8857288 22.32679 3.079228e-42 2.685292e-39 14.641737
ADAM11 124.55762 1.3794780 22.23870 4.373923e-42 3.575955e-39 13.827542
DCX 132.88650 2.6276693 22.05566 9.095757e-42 6.998918e-39 23.516234
GAP43 118.28617 4.5145804 21.91289 1.614457e-41 1.173262e-38 31.393742
OLFM1 119.02393 2.7883832 21.67892 4.155562e-41 2.860995e-38 21.073905
NEFL 120.25644 3.6652260 21.59438 5.857186e-41 3.830892e-38 26.908973
RGS10 148.12027 3.1825479 21.53906 7.335332e-41 4.569213e-38 29.335529
CRMP1 109.67937 3.7555245 21.35171 1.576082e-40 9.371237e-38 24.239414
EBF1 144.29651 1.4468041 21.06943 5.029074e-40 2.860231e-37 18.564648
NEUROD1 175.24574 1.6536858 20.74053 1.967234e-39 1.072224e-36 25.563619
TAGLN3 145.55961 3.8492674 20.61828 3.277108e-39 1.714714e-36 31.534294
CCER2 140.68671 0.6853025 20.30846 1.204213e-38 6.058578e-36 12.728506
CACNA1A 101.79501 3.3103615 20.27926 1.362211e-38 6.599661e-36 19.050942
RTN1 129.04158 3.1365014 20.18087 2.065233e-38 9.648325e-36 24.598575
P2RX3 123.10529 0.5999809 20.08437 3.109610e-38 1.402649e-35 9.016660
KLHL35 123.42616 2.1556421 19.95892 5.303060e-38 2.312311e-35 17.729740
RTN4RL2 131.68886 1.3617224 19.89776 6.884279e-38 2.904944e-35 14.281421
INA 107.09496 2.6686304 19.85483 8.270219e-38 3.380710e-35 16.603477
CDKN2D 83.54951 3.4783630 19.30821 8.721599e-37 3.457189e-34 15.527483
TUBB2B 64.95113 8.1127821 19.25257 1.110820e-36 4.273717e-34 38.000339
NEFM 139.27868 3.9768428 19.13767 1.832766e-36 6.849832e-34 29.103516
MAPK8IP1 97.12378 2.9322347 18.58273 2.106376e-35 7.653750e-33 14.703837
NTRK1 133.99739 0.7886271 18.46207 3.600259e-35 1.272838e-32 11.386000
SRRM4 121.29067 1.8829324 18.39436 4.867799e-35 1.675676e-32 14.086560
CD163L1 135.87117 0.3689427 18.11018 1.737174e-34 5.826661e-32 9.110318
EBF3 121.60265 1.2507997 17.88303 4.838159e-34 1.582199e-31 9.823714
SYP 116.31237 1.8687858 17.70193 1.099981e-33 3.509474e-31 12.485281
STMN3 101.55171 3.0624184 17.38022 4.780582e-33 1.488924e-30 15.420668
ISL1 121.27897 3.1288175 17.27599 7.716615e-33 2.347466e-30 20.639316
ONECUT2 123.48408 2.2247446 17.19986 1.095733e-32 3.257564e-30 15.772407
BASP1 66.78895 8.6309172 16.91702 4.056992e-32 1.179323e-29 36.939640
RBFOX3 108.79836 1.6880846 16.69109 1.162742e-31 3.306483e-29 9.291179
PPP1R27 121.14895 0.4451864 16.68133 1.216996e-31 3.387134e-29 5.652912
PRKG1 100.71415 2.9548203 16.59066 1.860503e-31 5.070259e-29 13.676546
TLCD3B 110.89862 1.7731339 16.42595 4.033215e-31 1.076704e-28 10.034900
CELF3 114.68471 1.2546600 16.33571 6.171335e-31 1.614545e-28 8.439524
$scHCL.hESC
logFC AveExpr t P.Value adj.P.Val B
SFRP2 88.47448 4.98786079 29.98283 3.932109e-54 5.143592e-50 69.069262
SOX2 86.08364 4.91896284 27.88873 3.996600e-51 2.613976e-47 66.678658
TUBB2B 39.55603 8.11278211 24.65245 3.994784e-46 1.741859e-42 75.749672
AIF1L 42.65084 4.43344569 22.87440 3.544932e-43 1.159281e-39 55.797943
CENPH 30.15820 5.89862049 22.35249 2.780067e-42 7.273211e-39 63.119952
PSIP1 28.33854 7.78625035 22.21049 4.895163e-42 1.067227e-38 70.201612
MXD4 -33.57232 6.83689314 -21.61983 5.281587e-41 9.869778e-38 63.084482
CDCA7L 39.90071 4.63147355 21.50205 8.528991e-41 1.394597e-37 55.178777
MCM3 28.16851 6.37609094 21.21454 2.766444e-40 4.020872e-37 62.385254
LEPROT -24.40255 6.67204168 -21.15175 3.581819e-40 4.685378e-37 61.428441
ADD2 57.19132 2.88840658 21.02830 5.959936e-40 7.087447e-37 45.678860
NUDT1 23.40843 6.25338182 20.94338 8.468967e-40 8.745369e-37 61.778780
PDZRN3 -67.72880 3.70102897 -20.93713 8.691216e-40 8.745369e-37 38.221765
GDF3 97.69448 1.59996404 20.81997 1.413393e-39 1.320614e-36 41.223821
MYOF -65.86157 3.52227670 -20.78941 1.604915e-39 1.321188e-36 35.768940
PARP1 23.27277 8.32541525 20.78776 1.616008e-39 1.321188e-36 67.123967
PAICS 21.93086 7.36177305 20.75609 1.843776e-39 1.418732e-36 65.174341
FOXB1 65.40662 1.17327676 20.38864 8.588984e-39 6.241806e-36 37.571795
SIPA1L2 -51.65337 6.17301564 -20.16709 2.189349e-38 1.484825e-35 52.672560
UNG 30.23598 5.29044745 20.15853 2.270201e-38 1.484825e-35 55.443573
DRAXIN 47.25994 3.38971110 19.86078 8.062656e-38 4.820975e-35 45.823462
DNAJC22 -94.03594 1.55877999 -19.85947 8.108054e-38 4.820975e-35 15.254184
EPSTI1 -73.70485 3.41671521 -19.79922 1.049219e-37 5.967322e-35 32.639722
CLRN3 -154.31051 -0.17763961 -19.67721 1.770897e-37 9.652127e-35 3.213731
EPS8L3 -149.66171 0.08879629 -19.64311 2.050587e-37 1.072949e-34 3.506354
SERPINA4 -162.01529 -0.25042397 -19.60148 2.453034e-37 1.234159e-34 3.027360
OIP5 31.95107 3.84347340 19.56390 2.884195e-37 1.397339e-34 46.674133
AURKB 35.28134 5.17861000 19.39822 5.902328e-37 2.757441e-34 52.676773
PRDM6 -93.68929 1.83879604 -19.22489 1.253043e-36 5.652086e-34 19.881953
SYTL5 -98.93367 2.24499018 -18.96092 3.972123e-36 1.731978e-33 18.278283
WNT11 -86.64629 2.08933026 -18.94104 4.334242e-36 1.828910e-33 21.444003
GLTPD2 -111.87363 0.85038922 -18.84462 6.621945e-36 2.706927e-33 8.194169
HELLS 37.67357 6.54538281 18.80437 7.906349e-36 3.134029e-33 57.218815
ALPL 47.41466 3.83780508 18.79601 8.203432e-36 3.156150e-33 44.644139
ANKS4B -130.71037 0.24721118 -18.72839 1.105523e-35 4.131812e-33 4.758401
GHR -80.55263 1.73281284 -18.63844 1.645607e-35 5.979497e-33 20.920098
DEK 21.12288 8.03922184 18.61120 1.856593e-35 6.563810e-33 59.678735
TM4SF5 -156.49140 -0.14786913 -18.57943 2.137395e-35 7.357701e-33 2.998603
F7 -65.00254 1.40758071 -18.51683 2.822126e-35 9.465699e-33 20.417254
SULT2A1 -147.01915 0.03410951 -18.43279 4.101657e-35 1.341344e-32 3.396117
POLR3G 75.99046 3.01002572 18.40034 4.739639e-35 1.512176e-32 42.203558
HP -134.03382 0.76214284 -18.39385 4.878817e-35 1.519519e-32 5.974937
SMLR1 -109.29633 1.18969314 -18.38294 5.122131e-35 1.558200e-32 10.182044
POU5F1 95.04689 1.07198687 18.36799 5.475388e-35 1.627808e-32 35.375326
HNF4A -113.17626 1.21622595 -18.35007 5.931185e-35 1.724129e-32 11.934774
CHAF1A 29.48863 5.39328572 18.33970 6.212265e-35 1.766579e-32 50.978346
MYLK3 -79.76192 1.47854340 -18.32711 6.571658e-35 1.829018e-32 18.850828
LHX1 -132.32825 0.18853835 -18.31336 6.988050e-35 1.904389e-32 4.296949
MYRF -63.06571 3.18296951 -18.29571 7.561781e-35 1.978685e-32 31.527621
CDCA5 33.53810 4.23890971 18.29567 7.563202e-35 1.978685e-32 45.848079checking for top DE markers from Cao and descartes browser
#metanephric: EYA1, UNCX, LRP2, NKAIN3, TMEM132D
tab<- topTable(efit, coef=14, sort.by="P", n=2000)
tab[rownames(tab) %in% c("EYA1", "UNCX", "LRP2", "NKAIN3", "TMEM132D", "IGF2", "AUTS2", "DACH1", "ROBO2", "H19", "PTPRG", "ERBB4", "NCAM", "FZD7", "SIX2", "OSR1", "WT1", "PAX2", "MEOX1", "MEIS1", "MEIS2"),] logFC AveExpr t P.Value adj.P.Val B
IGF2 132.18863 3.400974 17.139198 1.449645e-32 3.577889e-30 30.2514297
AUTS2 -37.07923 4.939878 -5.843750 5.496029e-08 1.018322e-06 -1.7166855
ERBB4 54.65935 3.671931 5.325014 5.555282e-07 8.808320e-06 0.6774518
ROBO2 -40.11892 4.068405 -3.298405 1.317749e-03 8.683867e-03 -3.8271393FeaturePlot(merged, features=c("EYA1", "UNCX", "LRP2", "NKAIN3", "TMEM132D"))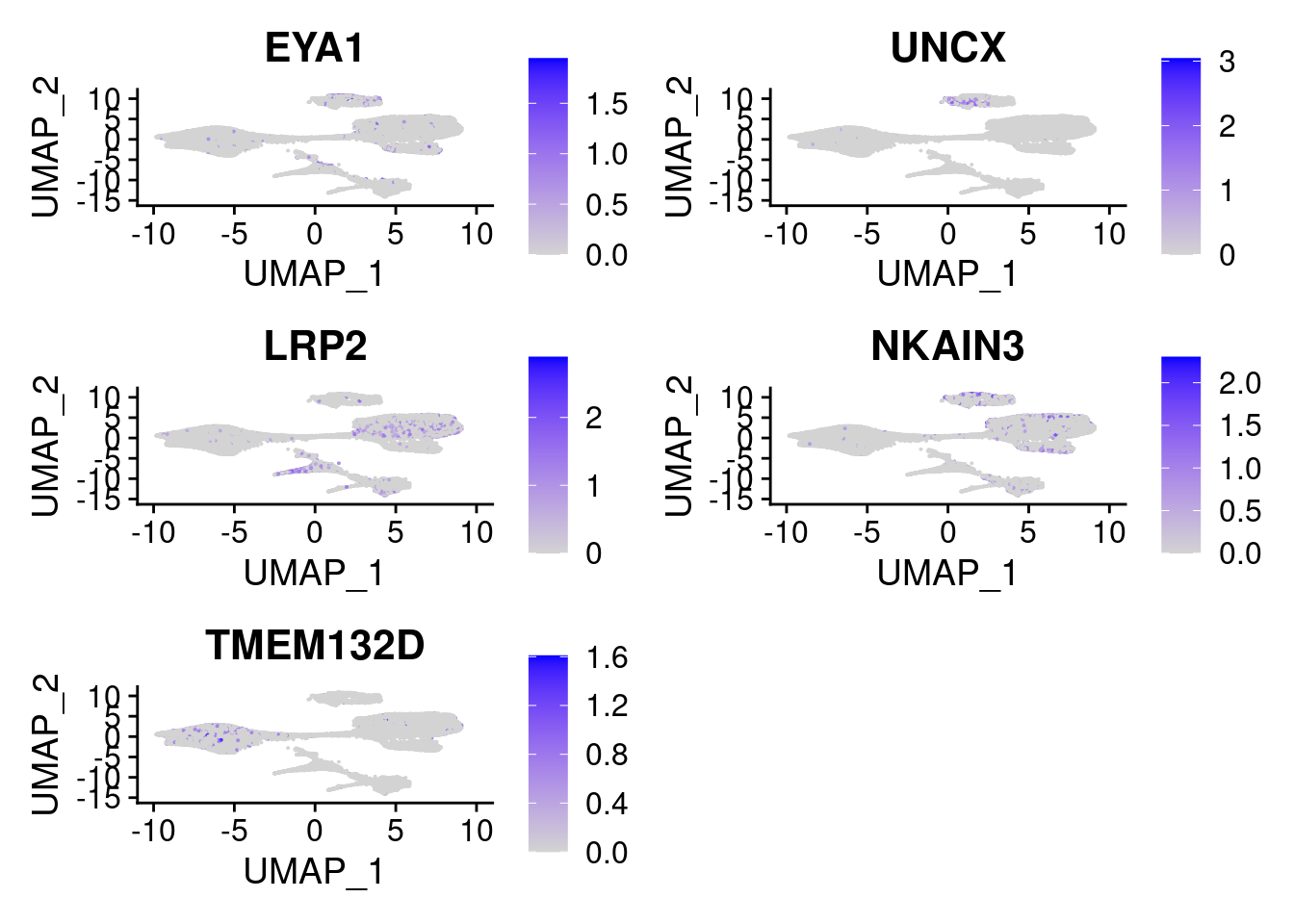
#hepatoblasts: SLC2A25, ACSS2, ASSI. SLC2A25 and ASS1 are not in our data set
tab<- topTable(efit, coef=8, sort.by="P", n=2000)
tab[rownames(tab) %in% c("SLC2A25", "ACSS2", "ASSI", "DLK1", "KRT18", "KRT8"),] logFC AveExpr t P.Value adj.P.Val B
ACSS2 62.36734 3.254362 16.01529 2.817248e-30 1.349905e-28 19.19883vol<- topTable(efit, coef=8, n=nrow(fit))
labsig<- vol[rownames(vol) %in% c("AFP", "FGB", "ACSS2"),]
labsiggenes<- rownames(labsig)
thresh<- vol$adj.P.Val < 0.05
vol<-cbind(vol, thresh)
b<- ggplot(vol, aes(x=logFC, y= -log10(adj.P.Val))) +
geom_point(aes(colour=thresh), show.legend = FALSE) +
scale_colour_manual(values = c("TRUE" = "red", "FALSE" = "black")) +
geom_text(data=labsig, aes(label=labsiggenes))
b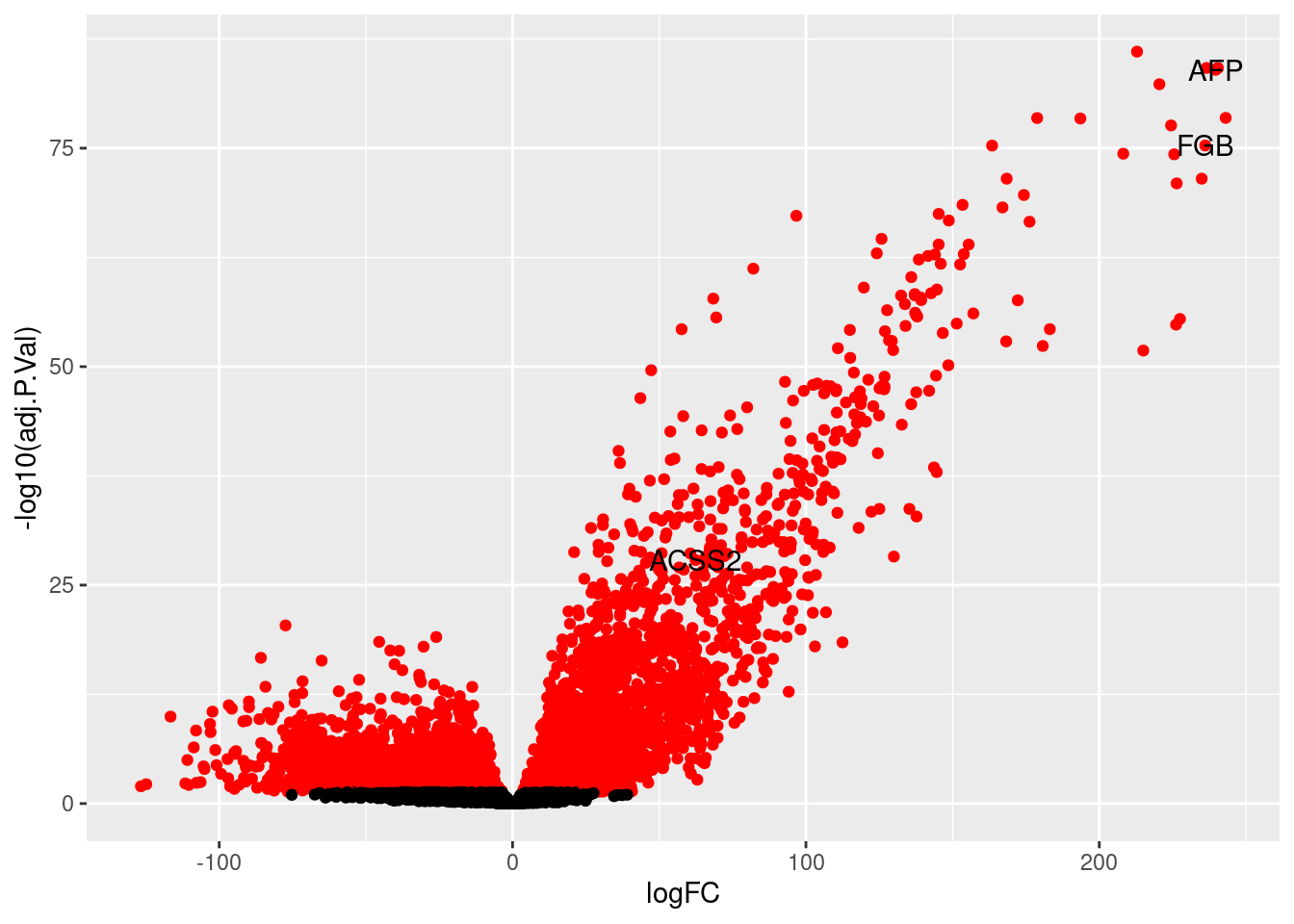
png(file= "/project2/gilad/katie/Pilot_HumanEBs/Embryoid_Body_Pilot_Workflowr/output/figs/Supp_RefAnnDE_hepatoblast.png", width= 6, height=6, units= "in", res= 1080)
b
dev.off()#amacrine: MEIS2
tab<- topTable(efit, coef=1, sort.by="P", n=2000)
tab[rownames(tab) %in% c("MEIS2"),][1] logFC AveExpr t P.Value adj.P.Val B
<0 rows> (or 0-length row.names)mesothelial
vol<- topTable(efit, coef=13, n=nrow(fit))
labsig<- vol[rownames(vol) %in% c("NID2", "COL6A3", "COL1A1", "COL3A1", "COL6A1"),]
labsiggenes<- rownames(labsig)
thresh<- vol$adj.P.Val < 0.05
vol<-cbind(vol, thresh)
p<- ggplot(vol, aes(x=logFC, y= -log10(adj.P.Val))) +
geom_point(aes(colour=thresh), show.legend = FALSE) +
scale_colour_manual(values = c("TRUE" = "red", "FALSE" = "black")) +
geom_text(data=labsig, aes(label=labsiggenes))
p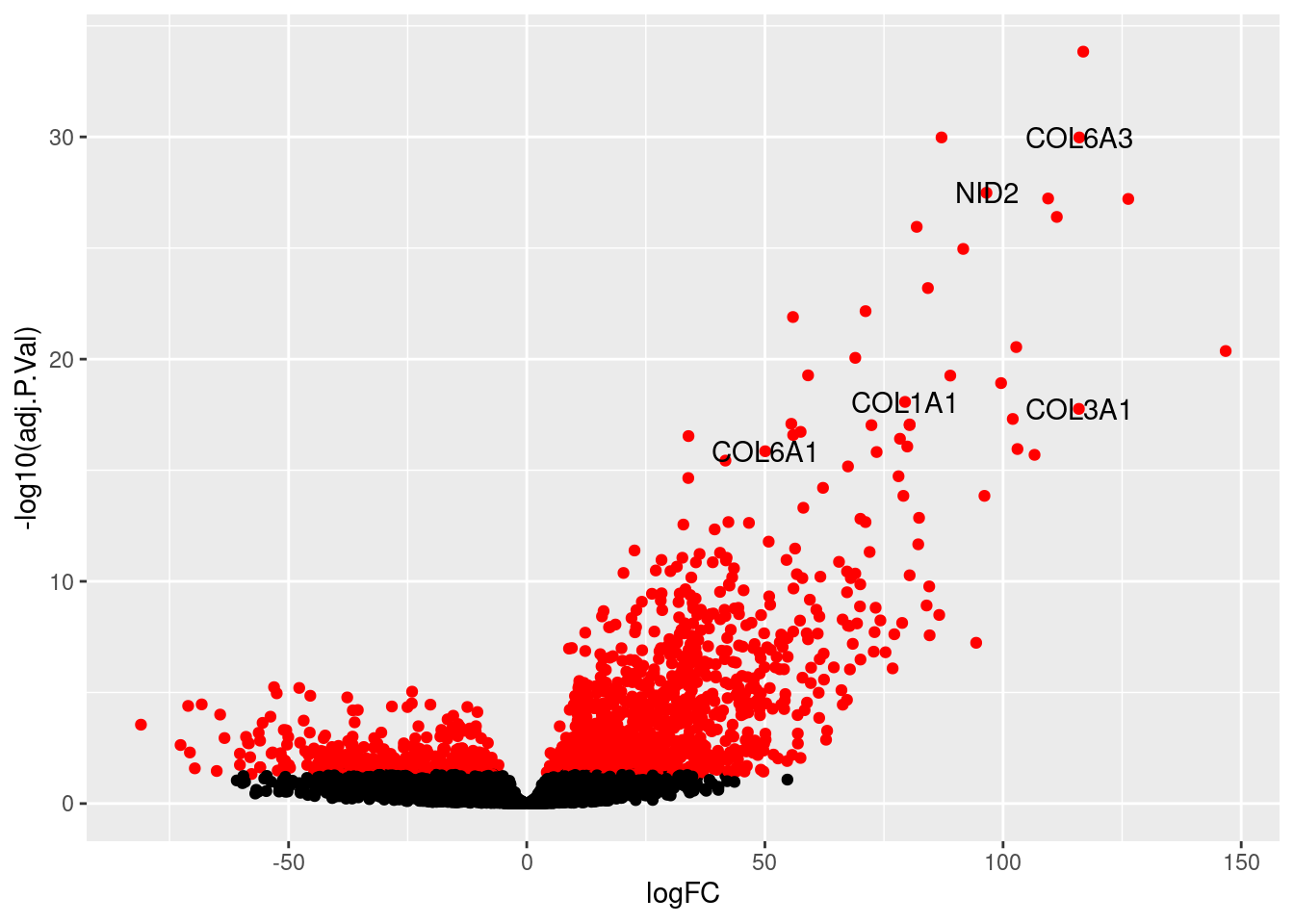
png(file= "/project2/gilad/katie/Pilot_HumanEBs/Embryoid_Body_Pilot_Workflowr/output/figs/Supp_RefAnnDE_mesothelial.png", width= 6, height=6, units= "in", res= 1080)
p
dev.off()sessionInfo()R version 3.6.1 (2019-07-05)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Scientific Linux 7.4 (Nitrogen)
Matrix products: default
BLAS/LAPACK: /software/openblas-0.2.19-el7-x86_64/lib/libopenblas_haswellp-r0.2.19.so
locale:
[1] C
attached base packages:
[1] parallel stats4 stats graphics grDevices utils datasets
[8] methods base
other attached packages:
[1] scater_1.14.6 ggplot2_3.3.3
[3] SingleCellExperiment_1.8.0 SummarizedExperiment_1.16.1
[5] DelayedArray_0.12.3 BiocParallel_1.20.1
[7] matrixStats_0.57.0 Biobase_2.46.0
[9] GenomicRanges_1.38.0 GenomeInfoDb_1.22.1
[11] IRanges_2.20.2 S4Vectors_0.24.4
[13] BiocGenerics_0.32.0 dplyr_1.0.2
[15] Matrix_1.2-18 edgeR_3.28.1
[17] limma_3.42.2 Seurat_3.2.0
[19] workflowr_1.6.2
loaded via a namespace (and not attached):
[1] plyr_1.8.6 igraph_1.2.6 lazyeval_0.2.2
[4] splines_3.6.1 listenv_0.8.0 digest_0.6.27
[7] htmltools_0.5.0 viridis_0.5.1 gdata_2.18.0
[10] magrittr_2.0.1 tensor_1.5 cluster_2.1.0
[13] ROCR_1.0-7 globals_0.12.5 colorspace_2.0-0
[16] rappdirs_0.3.3 ggrepel_0.9.0 xfun_0.16
[19] crayon_1.3.4 RCurl_1.98-1.2 jsonlite_1.7.2
[22] spatstat_1.64-1 spatstat.data_1.4-3 survival_3.2-3
[25] zoo_1.8-8 ape_5.4-1 glue_1.4.2
[28] polyclip_1.10-0 gtable_0.3.0 zlibbioc_1.32.0
[31] XVector_0.26.0 leiden_0.3.3 BiocSingular_1.2.2
[34] future.apply_1.6.0 abind_1.4-5 scales_1.1.1
[37] miniUI_0.1.1.1 Rcpp_1.0.6 viridisLite_0.3.0
[40] xtable_1.8-4 reticulate_1.20 rsvd_1.0.3
[43] htmlwidgets_1.5.1 httr_1.4.2 gplots_3.0.4
[46] RColorBrewer_1.1-2 ellipsis_0.3.1 ica_1.0-2
[49] farver_2.0.3 pkgconfig_2.0.3 uwot_0.1.10
[52] deldir_0.1-28 locfit_1.5-9.4 labeling_0.4.2
[55] tidyselect_1.1.0 rlang_0.4.10 reshape2_1.4.4
[58] later_1.1.0.1 munsell_0.5.0 tools_3.6.1
[61] generics_0.1.0 ggridges_0.5.2 evaluate_0.14
[64] stringr_1.4.0 fastmap_1.0.1 yaml_2.2.1
[67] goftest_1.2-2 npsurv_0.4-0 knitr_1.29
[70] fs_1.4.2 fitdistrplus_1.0-14 caTools_1.18.0
[73] purrr_0.3.4 RANN_2.6.1 pbapply_1.4-2
[76] future_1.18.0 nlme_3.1-140 mime_0.9
[79] compiler_3.6.1 beeswarm_0.2.3 plotly_4.9.2.1
[82] png_0.1-7 lsei_1.2-0 spatstat.utils_1.17-0
[85] tibble_3.0.4 stringi_1.5.3 highr_0.8
[88] lattice_0.20-38 vctrs_0.3.6 pillar_1.4.7
[91] lifecycle_0.2.0 lmtest_0.9-37 RcppAnnoy_0.0.18
[94] BiocNeighbors_1.4.2 data.table_1.13.4 cowplot_1.1.1
[97] bitops_1.0-6 irlba_2.3.3 httpuv_1.5.4
[100] patchwork_1.1.1 R6_2.5.0 promises_1.1.1
[103] KernSmooth_2.23-15 gridExtra_2.3 vipor_0.4.5
[106] codetools_0.2-16 MASS_7.3-51.4 gtools_3.8.2
[109] rprojroot_2.0.2 withr_2.4.2 sctransform_0.2.1
[112] GenomeInfoDbData_1.2.2 mgcv_1.8-28 grid_3.6.1
[115] rpart_4.1-15 tidyr_1.1.0 DelayedMatrixStats_1.8.0
[118] rmarkdown_2.3 Rtsne_0.15 git2r_0.26.1
[121] shiny_1.5.0 ggbeeswarm_0.6.0
sessionInfo()R version 3.6.1 (2019-07-05)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Scientific Linux 7.4 (Nitrogen)
Matrix products: default
BLAS/LAPACK: /software/openblas-0.2.19-el7-x86_64/lib/libopenblas_haswellp-r0.2.19.so
locale:
[1] C
attached base packages:
[1] parallel stats4 stats graphics grDevices utils datasets
[8] methods base
other attached packages:
[1] scater_1.14.6 ggplot2_3.3.3
[3] SingleCellExperiment_1.8.0 SummarizedExperiment_1.16.1
[5] DelayedArray_0.12.3 BiocParallel_1.20.1
[7] matrixStats_0.57.0 Biobase_2.46.0
[9] GenomicRanges_1.38.0 GenomeInfoDb_1.22.1
[11] IRanges_2.20.2 S4Vectors_0.24.4
[13] BiocGenerics_0.32.0 dplyr_1.0.2
[15] Matrix_1.2-18 edgeR_3.28.1
[17] limma_3.42.2 Seurat_3.2.0
[19] workflowr_1.6.2
loaded via a namespace (and not attached):
[1] plyr_1.8.6 igraph_1.2.6 lazyeval_0.2.2
[4] splines_3.6.1 listenv_0.8.0 digest_0.6.27
[7] htmltools_0.5.0 viridis_0.5.1 gdata_2.18.0
[10] magrittr_2.0.1 tensor_1.5 cluster_2.1.0
[13] ROCR_1.0-7 globals_0.12.5 colorspace_2.0-0
[16] rappdirs_0.3.3 ggrepel_0.9.0 xfun_0.16
[19] crayon_1.3.4 RCurl_1.98-1.2 jsonlite_1.7.2
[22] spatstat_1.64-1 spatstat.data_1.4-3 survival_3.2-3
[25] zoo_1.8-8 ape_5.4-1 glue_1.4.2
[28] polyclip_1.10-0 gtable_0.3.0 zlibbioc_1.32.0
[31] XVector_0.26.0 leiden_0.3.3 BiocSingular_1.2.2
[34] future.apply_1.6.0 abind_1.4-5 scales_1.1.1
[37] miniUI_0.1.1.1 Rcpp_1.0.6 viridisLite_0.3.0
[40] xtable_1.8-4 reticulate_1.20 rsvd_1.0.3
[43] htmlwidgets_1.5.1 httr_1.4.2 gplots_3.0.4
[46] RColorBrewer_1.1-2 ellipsis_0.3.1 ica_1.0-2
[49] farver_2.0.3 pkgconfig_2.0.3 uwot_0.1.10
[52] deldir_0.1-28 locfit_1.5-9.4 labeling_0.4.2
[55] tidyselect_1.1.0 rlang_0.4.10 reshape2_1.4.4
[58] later_1.1.0.1 munsell_0.5.0 tools_3.6.1
[61] generics_0.1.0 ggridges_0.5.2 evaluate_0.14
[64] stringr_1.4.0 fastmap_1.0.1 yaml_2.2.1
[67] goftest_1.2-2 npsurv_0.4-0 knitr_1.29
[70] fs_1.4.2 fitdistrplus_1.0-14 caTools_1.18.0
[73] purrr_0.3.4 RANN_2.6.1 pbapply_1.4-2
[76] future_1.18.0 nlme_3.1-140 mime_0.9
[79] compiler_3.6.1 beeswarm_0.2.3 plotly_4.9.2.1
[82] png_0.1-7 lsei_1.2-0 spatstat.utils_1.17-0
[85] tibble_3.0.4 stringi_1.5.3 highr_0.8
[88] lattice_0.20-38 vctrs_0.3.6 pillar_1.4.7
[91] lifecycle_0.2.0 lmtest_0.9-37 RcppAnnoy_0.0.18
[94] BiocNeighbors_1.4.2 data.table_1.13.4 cowplot_1.1.1
[97] bitops_1.0-6 irlba_2.3.3 httpuv_1.5.4
[100] patchwork_1.1.1 R6_2.5.0 promises_1.1.1
[103] KernSmooth_2.23-15 gridExtra_2.3 vipor_0.4.5
[106] codetools_0.2-16 MASS_7.3-51.4 gtools_3.8.2
[109] rprojroot_2.0.2 withr_2.4.2 sctransform_0.2.1
[112] GenomeInfoDbData_1.2.2 mgcv_1.8-28 grid_3.6.1
[115] rpart_4.1-15 tidyr_1.1.0 DelayedMatrixStats_1.8.0
[118] rmarkdown_2.3 Rtsne_0.15 git2r_0.26.1
[121] shiny_1.5.0 ggbeeswarm_0.6.0